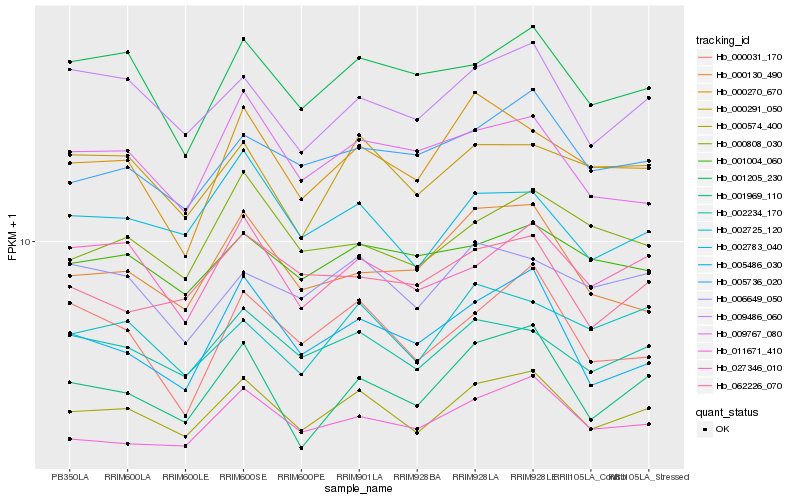
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_400 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15930 [Jatropha curcas] |
| 2 |
Hb_002234_170 |
0.0682049686 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002783_040 |
0.0751200133 |
transcription factor |
TF Family: IWS1 |
- |
| 4 |
Hb_009486_060 |
0.0770226285 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 5 |
Hb_027346_010 |
0.0785169668 |
- |
- |
- |
| 6 |
Hb_001205_230 |
0.0806935068 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 7 |
Hb_011671_410 |
0.0818083604 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 8 |
Hb_005486_030 |
0.0819445749 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 9 |
Hb_000270_670 |
0.0826279795 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 10 |
Hb_000291_050 |
0.0838527301 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002725_120 |
0.084392596 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005736_020 |
0.0843939259 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 13 |
Hb_001969_110 |
0.0850916537 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g15510, chloroplastic [Jatropha curcas] |
| 14 |
Hb_006649_050 |
0.0871864595 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 15 |
Hb_009767_080 |
0.0880897358 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 16 |
Hb_001004_060 |
0.0890247969 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 17 |
Hb_000130_490 |
0.0908196742 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000808_030 |
0.0908268245 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 19 |
Hb_062226_070 |
0.0910838318 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000031_170 |
0.091242906 |
- |
- |
PREDICTED: abnormal long morphology protein 1 isoform X1 [Jatropha curcas] |