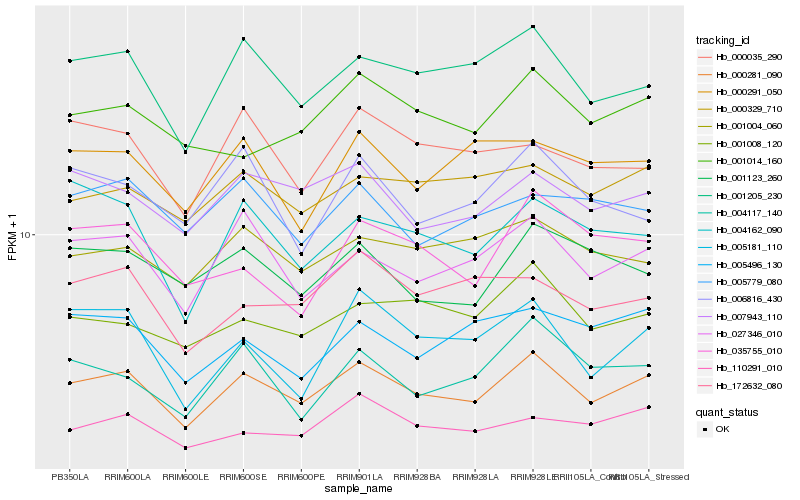
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000281_090 |
0.0 |
transcription factor |
TF Family: SNF2 |
hypothetical protein PRUPE_ppa000063mg [Prunus persica] |
| 2 |
Hb_001205_230 |
0.0667754765 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 3 |
Hb_027346_010 |
0.0723058041 |
- |
- |
- |
| 4 |
Hb_007943_110 |
0.0808304259 |
- |
- |
PREDICTED: basic salivary proline-rich protein 3 [Jatropha curcas] |
| 5 |
Hb_001008_120 |
0.0882663745 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 6 |
Hb_001004_060 |
0.0900439666 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 7 |
Hb_004162_090 |
0.0903926427 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 8 |
Hb_172632_080 |
0.0911126036 |
- |
- |
PREDICTED: uncharacterized protein LOC105141138 isoform X1 [Populus euphratica] |
| 9 |
Hb_110291_010 |
0.0911442013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000329_710 |
0.0912391404 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001014_160 |
0.0921712513 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 12 |
Hb_001123_260 |
0.0923379063 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 13 |
Hb_004117_140 |
0.0943426573 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g16710, mitochondrial [Populus euphratica] |
| 14 |
Hb_005181_110 |
0.0950304683 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 15 |
Hb_000291_050 |
0.0953053193 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000035_290 |
0.0957281632 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 17 |
Hb_005496_130 |
0.0958833794 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_005779_080 |
0.0959629515 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006816_430 |
0.0969352815 |
- |
- |
PREDICTED: topless-related protein 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_035755_010 |
0.0979704155 |
- |
- |
unknown [Populus trichocarpa] |