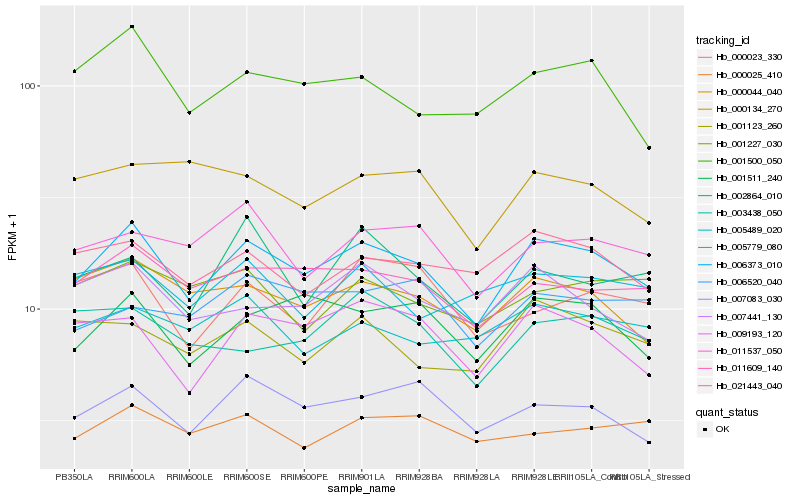
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006373_010 |
0.0 |
- |
- |
Potassium transporter 11 [Gossypium arboreum] |
| 2 |
Hb_011609_140 |
0.08109338 |
- |
- |
hypothetical protein JCGZ_12974 [Jatropha curcas] |
| 3 |
Hb_001511_240 |
0.0813294773 |
- |
- |
PREDICTED: uncharacterized protein LOC105133574 isoform X3 [Populus euphratica] |
| 4 |
Hb_009193_120 |
0.0839181785 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 5 |
Hb_007083_030 |
0.0849524224 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 6 |
Hb_000044_040 |
0.0866696428 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 7 |
Hb_021443_040 |
0.0910788807 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 8 |
Hb_007441_130 |
0.0913003856 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001500_050 |
0.0935967163 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 10 |
Hb_011537_050 |
0.0940237274 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001123_260 |
0.0947280795 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 12 |
Hb_005489_020 |
0.0975450388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000025_410 |
0.0995410959 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g01110-like [Populus euphratica] |
| 14 |
Hb_005779_080 |
0.1024646659 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006520_040 |
0.1030254548 |
- |
- |
PREDICTED: uncharacterized protein LOC105641714 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003438_050 |
0.1036439436 |
- |
- |
PREDICTED: rab3 GTPase-activating protein catalytic subunit isoform X2 [Jatropha curcas] |
| 17 |
Hb_001227_030 |
0.1038560463 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002864_010 |
0.1048643242 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 19 |
Hb_000134_270 |
0.1048774736 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 20 |
Hb_000023_330 |
0.104940953 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |