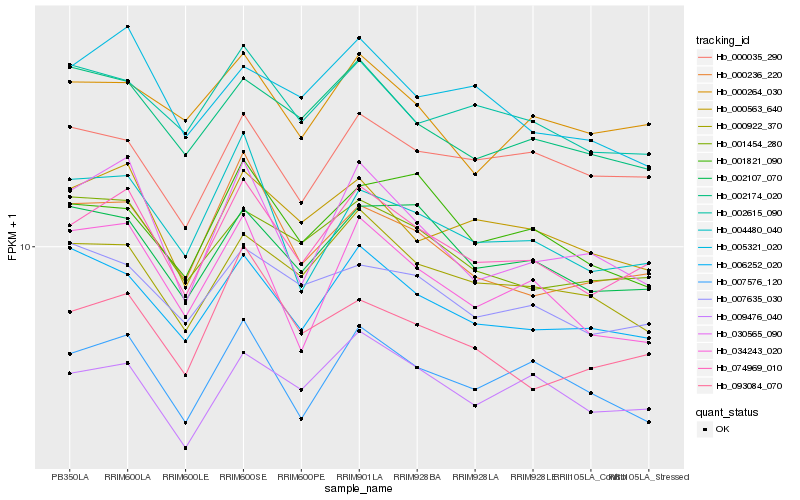
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074969_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23339 [Jatropha curcas] |
| 2 |
Hb_000563_640 |
0.0736648397 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 3 |
Hb_002107_070 |
0.0761858325 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 4 |
Hb_000922_370 |
0.0771021661 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 5 |
Hb_000236_220 |
0.0778428963 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 6 |
Hb_001454_280 |
0.0778505176 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 7 |
Hb_006252_020 |
0.0778860145 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_009476_040 |
0.0789061579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002615_090 |
0.0803032594 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007576_120 |
0.0819064534 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 11 |
Hb_030565_090 |
0.0821167679 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 12 |
Hb_002174_020 |
0.0825057035 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_000264_030 |
0.0834013744 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 14 |
Hb_004480_040 |
0.0842845891 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_093084_070 |
0.0846362978 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 16 |
Hb_000035_290 |
0.0848146165 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 17 |
Hb_007635_030 |
0.0856056465 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_005321_020 |
0.0869744606 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 19 |
Hb_001821_090 |
0.088450864 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 20 |
Hb_034243_020 |
0.0884698052 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |