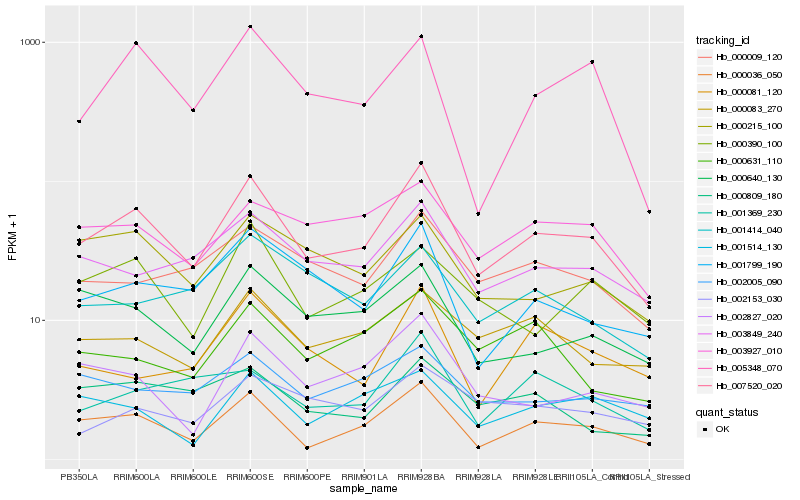
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007520_020 |
0.0 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
1-deoxy-D-xylulose-5-phosphate reductoisomerase [Hevea brasiliensis] |
| 2 |
Hb_000036_050 |
0.0963486452 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6-like [Camelina sativa] |
| 3 |
Hb_003849_240 |
0.1351190435 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001799_190 |
0.1424831317 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 5 |
Hb_000009_120 |
0.1435112097 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 6 |
Hb_000640_130 |
0.1470719692 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_003927_010 |
0.1475140055 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_000083_270 |
0.1482884016 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000215_100 |
0.1490585889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005348_070 |
0.150127242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002827_020 |
0.1533837107 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 12 |
Hb_000390_100 |
0.1536744546 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 13 |
Hb_002005_090 |
0.1547475161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001369_230 |
0.1591950443 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 15 |
Hb_000809_180 |
0.1624033667 |
- |
- |
- |
| 16 |
Hb_001414_040 |
0.1630879865 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000631_110 |
0.1654253703 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 18 |
Hb_002153_030 |
0.1658133962 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 19 |
Hb_000081_120 |
0.1662228786 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 20 |
Hb_001514_130 |
0.1678695587 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |