| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
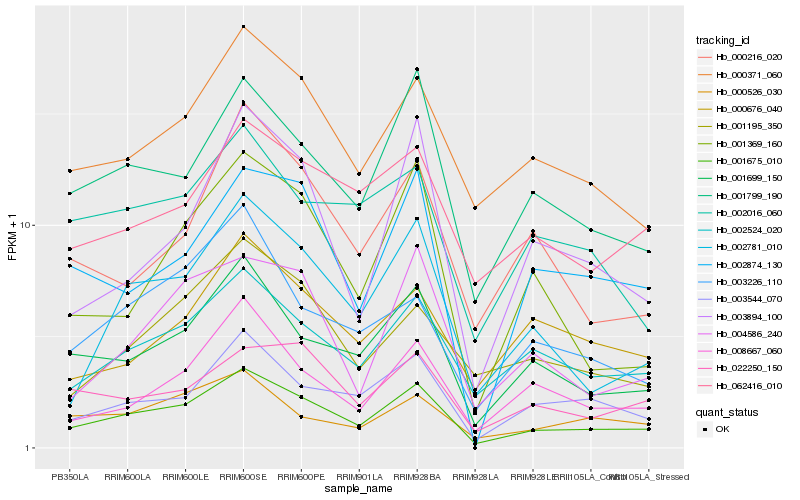
Hb_001675_010 |
0.0 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 2 |
Hb_001699_150 |
0.102996166 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 3 |
Hb_002524_020 |
0.1068247026 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 4 |
Hb_001799_190 |
0.1084169104 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 5 |
Hb_000371_060 |
0.1149431099 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 6 |
Hb_003544_070 |
0.115358949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002781_010 |
0.1236392832 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_008667_060 |
0.1305347453 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 9 |
Hb_062416_010 |
0.1355695415 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 10 |
Hb_022250_150 |
0.1378350169 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 11 |
Hb_000526_030 |
0.1385252705 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 12 |
Hb_001195_350 |
0.1392888711 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 13 |
Hb_004586_240 |
0.1403295657 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 14 |
Hb_003226_110 |
0.14097978 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 15 |
Hb_002874_130 |
0.1413623824 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 16 |
Hb_002016_060 |
0.1428798644 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 17 |
Hb_001369_160 |
0.1463263785 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 18 |
Hb_000216_020 |
0.1478171466 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 19 |
Hb_000676_040 |
0.1479929404 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 20 |
Hb_003894_100 |
0.1502805009 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |