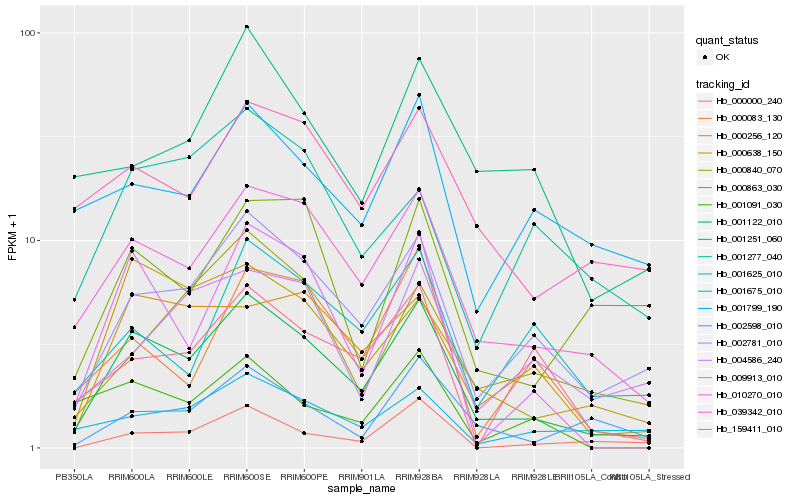
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001122_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 2 |
Hb_010270_010 |
0.1174331074 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_002781_010 |
0.1295524558 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000638_150 |
0.1609991935 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 5 |
Hb_009913_010 |
0.1693783943 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000256_120 |
0.1744932694 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 7 |
Hb_039342_010 |
0.1897090203 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 8 |
Hb_000083_130 |
0.1951093568 |
- |
- |
hypothetical protein RCOM_0884570 [Ricinus communis] |
| 9 |
Hb_001675_010 |
0.1953709515 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 10 |
Hb_000840_070 |
0.1969708364 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002598_010 |
0.2021619103 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 12 |
Hb_001625_010 |
0.2025869303 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 13 |
Hb_004586_240 |
0.2043608348 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 14 |
Hb_001251_060 |
0.2046185411 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 15 |
Hb_159411_010 |
0.2095152753 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 16 |
Hb_001799_190 |
0.2118661728 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 17 |
Hb_001277_040 |
0.2118872207 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 18 |
Hb_000863_030 |
0.2123768239 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 19 |
Hb_001091_030 |
0.2124938101 |
- |
- |
hypothetical protein AALP_AA1G173500 [Arabis alpina] |
| 20 |
Hb_000000_240 |
0.2147917543 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |