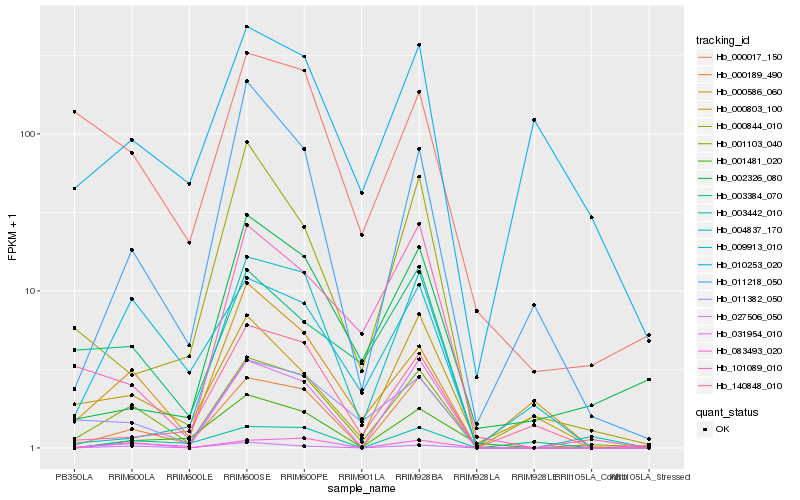
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001103_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 2 |
Hb_000189_490 |
0.1416712053 |
- |
- |
Sulfate/thiosulfate import ATP-binding protein cysA [Theobroma cacao] |
| 3 |
Hb_011382_050 |
0.1849908597 |
- |
- |
hypothetical protein PRUPE_ppa025935mg [Prunus persica] |
| 4 |
Hb_003442_010 |
0.1894055227 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 4-like isoform X2 [Citrus sinensis] |
| 5 |
Hb_083493_020 |
0.1994839966 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Vitis vinifera] |
| 6 |
Hb_027506_050 |
0.2050022358 |
- |
- |
- |
| 7 |
Hb_009913_010 |
0.2105247823 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000586_060 |
0.2159240474 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 9 |
Hb_011218_050 |
0.2164089585 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_003384_070 |
0.2183857396 |
- |
- |
PREDICTED: methylesterase 17-like [Populus euphratica] |
| 11 |
Hb_101089_010 |
0.2264252854 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 12 |
Hb_001481_020 |
0.2274136547 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase V.9-like [Jatropha curcas] |
| 13 |
Hb_000803_100 |
0.2292457782 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 14 |
Hb_000017_150 |
0.230785673 |
- |
- |
hypothetical protein POPTR_0001s09780g [Populus trichocarpa] |
| 15 |
Hb_004837_170 |
0.2334436612 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 16 |
Hb_031954_010 |
0.2355013941 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_140848_010 |
0.2361956098 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 18 |
Hb_002326_080 |
0.2372663498 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 19 |
Hb_000844_010 |
0.2384268827 |
- |
- |
PREDICTED: lysine histidine transporter-like 8 [Jatropha curcas] |
| 20 |
Hb_010253_020 |
0.2400726625 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |