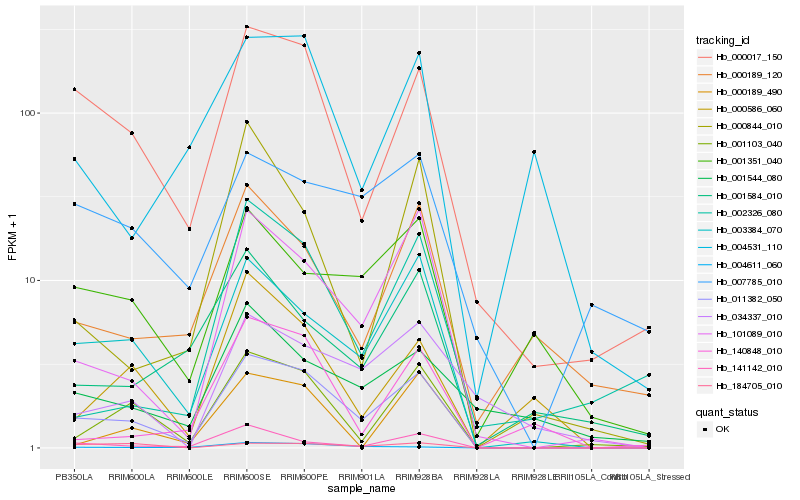
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011382_050 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa025935mg [Prunus persica] |
| 2 |
Hb_003384_070 |
0.1480858848 |
- |
- |
PREDICTED: methylesterase 17-like [Populus euphratica] |
| 3 |
Hb_000017_150 |
0.1665441762 |
- |
- |
hypothetical protein POPTR_0001s09780g [Populus trichocarpa] |
| 4 |
Hb_101089_010 |
0.1689809173 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 5 |
Hb_001103_040 |
0.1849908597 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 6 |
Hb_141142_010 |
0.196672461 |
- |
- |
hypothetical protein JCGZ_21320 [Jatropha curcas] |
| 7 |
Hb_004611_060 |
0.2278536734 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 8 |
Hb_001351_040 |
0.2322056087 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 2, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000586_060 |
0.2335735797 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 10 |
Hb_000189_490 |
0.2356891357 |
- |
- |
Sulfate/thiosulfate import ATP-binding protein cysA [Theobroma cacao] |
| 11 |
Hb_184705_010 |
0.2372894231 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000189_120 |
0.2373568854 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 13 |
Hb_000844_010 |
0.2411787132 |
- |
- |
PREDICTED: lysine histidine transporter-like 8 [Jatropha curcas] |
| 14 |
Hb_140848_010 |
0.2424082641 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 15 |
Hb_034337_010 |
0.2436687826 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 16 |
Hb_001584_010 |
0.2437172243 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 17 |
Hb_007785_010 |
0.2467097029 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 18 |
Hb_002326_080 |
0.2472322355 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 19 |
Hb_004531_110 |
0.2472909445 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 20 |
Hb_001544_080 |
0.24808479 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |