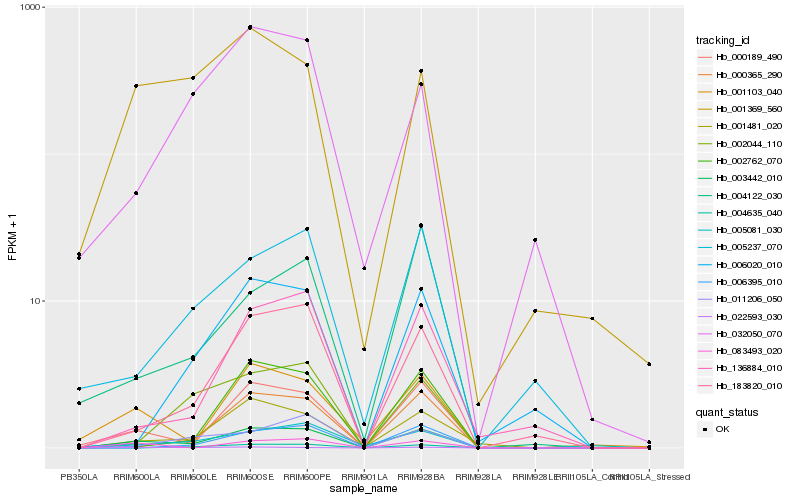
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003442_010 |
0.0 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 4-like isoform X2 [Citrus sinensis] |
| 2 |
Hb_000189_490 |
0.1442675138 |
- |
- |
Sulfate/thiosulfate import ATP-binding protein cysA [Theobroma cacao] |
| 3 |
Hb_001481_020 |
0.1672214849 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase V.9-like [Jatropha curcas] |
| 4 |
Hb_183820_010 |
0.1838386285 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 5 |
Hb_001103_040 |
0.1894055227 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 6 |
Hb_000365_290 |
0.2004764113 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_083493_020 |
0.2011796985 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Vitis vinifera] |
| 8 |
Hb_022593_030 |
0.2027725202 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 9 |
Hb_136884_010 |
0.210647824 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 10 |
Hb_005081_030 |
0.2149626168 |
- |
- |
hypothetical protein POPTR_0018s09200g [Populus trichocarpa] |
| 11 |
Hb_001369_560 |
0.2164721855 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 12 |
Hb_005237_070 |
0.2203742895 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 13 |
Hb_004635_040 |
0.2241375838 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 14 |
Hb_002044_110 |
0.2250645393 |
- |
- |
PREDICTED: probable carboxylesterase 5 isoform X1 [Populus euphratica] |
| 15 |
Hb_002762_070 |
0.2253799853 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 16 |
Hb_006020_010 |
0.232145078 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 17 |
Hb_006395_010 |
0.232688966 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 18 |
Hb_011206_050 |
0.2346716253 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 19 |
Hb_004122_030 |
0.2356348828 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 20 |
Hb_032050_070 |
0.2367662337 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |