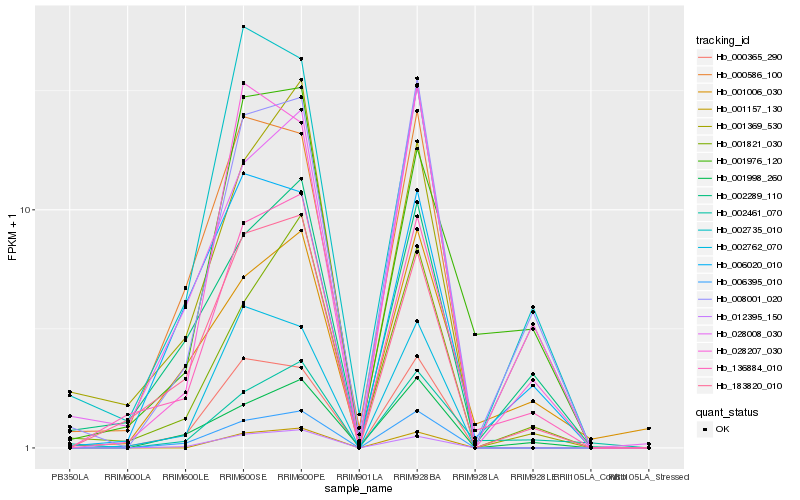
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_136884_010 |
0.0 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 2 |
Hb_183820_010 |
0.0892841232 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 3 |
Hb_002289_110 |
0.1318641184 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 4 |
Hb_001976_120 |
0.135399882 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_008001_020 |
0.1393461253 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 6 |
Hb_012395_150 |
0.1395156177 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 7 |
Hb_000365_290 |
0.1452978365 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_006395_010 |
0.1469141019 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 9 |
Hb_001369_530 |
0.1476113287 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 10 |
Hb_001821_030 |
0.1509302192 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 11 |
Hb_000586_100 |
0.1544461309 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002762_070 |
0.1560555056 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 13 |
Hb_006020_010 |
0.157623837 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 14 |
Hb_001998_260 |
0.1595414255 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_001006_030 |
0.1605682182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_028008_030 |
0.1637296582 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 17 |
Hb_002735_010 |
0.1639913966 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 44 [Prunus mume] |
| 18 |
Hb_002461_070 |
0.1660469092 |
- |
- |
gibberellin 2-oxidase, putative [Ricinus communis] |
| 19 |
Hb_028207_030 |
0.1671514801 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 20 |
Hb_001157_130 |
0.1686534213 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |