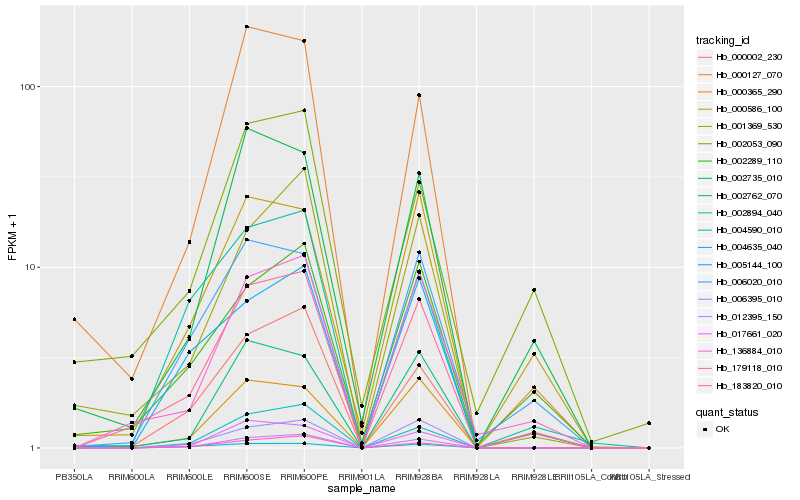
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183820_010 |
0.0 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 2 |
Hb_136884_010 |
0.0892841232 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 3 |
Hb_012395_150 |
0.1144140505 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 4 |
Hb_000002_230 |
0.1230993501 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 5 |
Hb_000365_290 |
0.1328457601 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_006020_010 |
0.1333166105 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 7 |
Hb_004590_010 |
0.1341344711 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 8 |
Hb_179118_010 |
0.1357234312 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_002289_110 |
0.1368523418 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 10 |
Hb_001369_530 |
0.1418608033 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 11 |
Hb_002735_010 |
0.1424432374 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 44 [Prunus mume] |
| 12 |
Hb_006395_010 |
0.143668824 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 13 |
Hb_017661_020 |
0.1469612859 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 14 |
Hb_002762_070 |
0.1471225022 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 15 |
Hb_000586_100 |
0.1476353498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004635_040 |
0.1491121169 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 17 |
Hb_000127_070 |
0.1496089642 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 18 |
Hb_005144_100 |
0.1512503955 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 19 |
Hb_002894_040 |
0.1547951154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002053_090 |
0.1580844429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |