| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
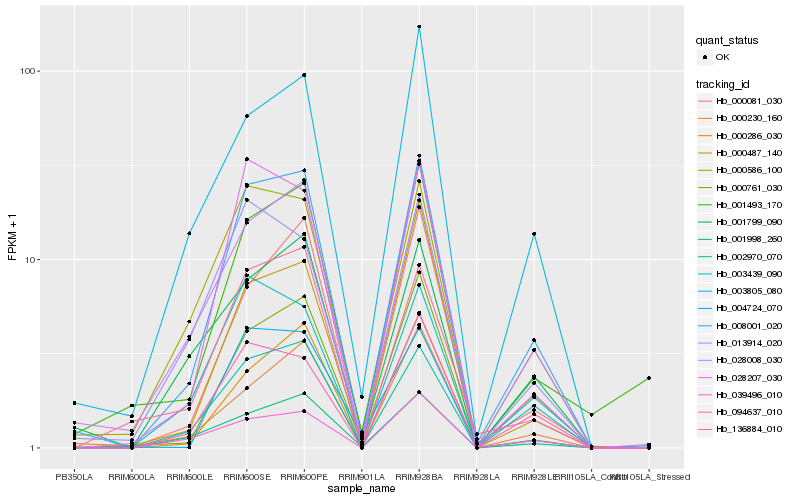
Hb_008001_020 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 2 |
Hb_028008_030 |
0.1089206207 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 3 |
Hb_000586_100 |
0.1179789341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_094637_010 |
0.1194941647 |
- |
- |
PREDICTED: uncharacterized protein LOC105642042 [Jatropha curcas] |
| 5 |
Hb_000286_030 |
0.1246370829 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 6 |
Hb_028207_030 |
0.1274065941 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 7 |
Hb_001998_260 |
0.1289193891 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_000761_030 |
0.1314719619 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 9 |
Hb_003439_090 |
0.1329890044 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000081_030 |
0.1375987561 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 11 |
Hb_001799_090 |
0.1392042809 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 12 |
Hb_136884_010 |
0.1393461253 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 13 |
Hb_000230_160 |
0.1396204788 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003805_080 |
0.1417879025 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 15 |
Hb_039496_010 |
0.1419852088 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 16 |
Hb_004724_070 |
0.1431027993 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000487_140 |
0.1450700149 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_001493_170 |
0.1469857375 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 19 |
Hb_013914_020 |
0.1470774821 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 20 |
Hb_002970_070 |
0.1521916605 |
- |
- |
multidrug/pheromone exporter protein [Hevea brasiliensis] |