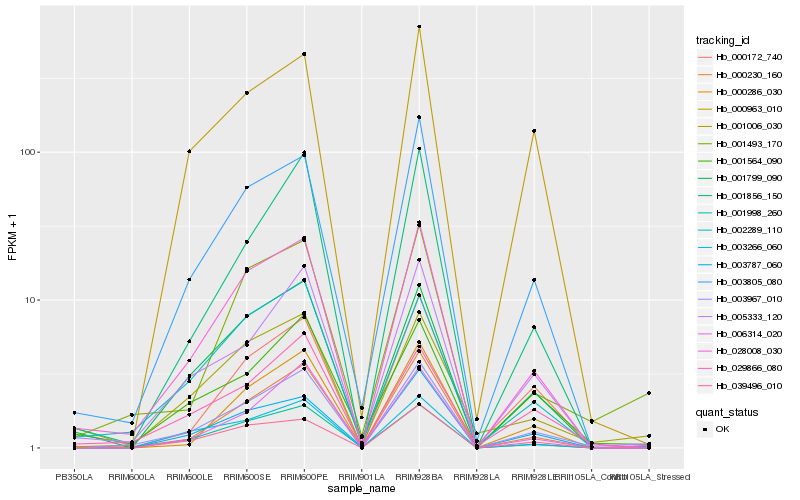
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003967_010 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_028008_030 |
0.106528579 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 3 |
Hb_000230_160 |
0.1185978444 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001006_030 |
0.1206846822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003266_060 |
0.1284839477 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 6 |
Hb_001493_170 |
0.1288362424 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 7 |
Hb_005333_120 |
0.1288397661 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 8 |
Hb_003805_080 |
0.1315404694 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 9 |
Hb_029866_080 |
0.1363072538 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 10 |
Hb_039496_010 |
0.1369300012 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 11 |
Hb_001998_260 |
0.1375731311 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_006314_020 |
0.1392428273 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 13 |
Hb_000963_010 |
0.1420616007 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 14 |
Hb_003787_060 |
0.1454391499 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 15 |
Hb_001856_150 |
0.1459923683 |
- |
- |
- |
| 16 |
Hb_001799_090 |
0.1466836424 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 17 |
Hb_001564_090 |
0.146704495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000172_740 |
0.1469631905 |
- |
- |
HXXXD-type acyl-transferase family protein, putative [Theobroma cacao] |
| 19 |
Hb_002289_110 |
0.1477619484 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 20 |
Hb_000286_030 |
0.1478361581 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |