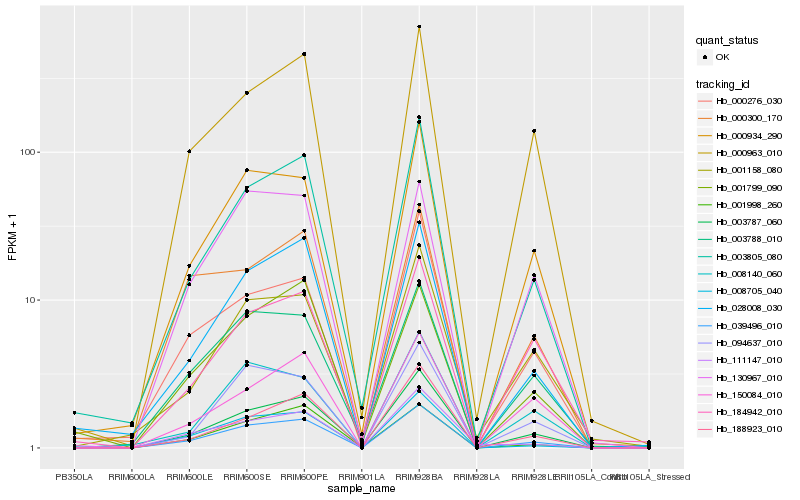
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039496_010 |
0.0 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 2 |
Hb_003787_060 |
0.0637923588 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 3 |
Hb_003805_080 |
0.0814941458 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 4 |
Hb_000934_290 |
0.0829446487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000963_010 |
0.0880636542 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 6 |
Hb_028008_030 |
0.0966626961 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 7 |
Hb_003788_010 |
0.0996298087 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 8 |
Hb_008705_040 |
0.1052270441 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 9 |
Hb_188923_010 |
0.114206913 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 10 |
Hb_094637_010 |
0.1151497642 |
- |
- |
PREDICTED: uncharacterized protein LOC105642042 [Jatropha curcas] |
| 11 |
Hb_001998_260 |
0.1170367963 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_150084_010 |
0.1233756192 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 13 |
Hb_000276_030 |
0.1238276148 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 14 |
Hb_130967_010 |
0.1239907596 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_111147_010 |
0.1260214631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001158_080 |
0.1268720224 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 17 |
Hb_184942_010 |
0.1283409682 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 18 |
Hb_008140_060 |
0.1292645443 |
- |
- |
Cytochrome P450, putative [Theobroma cacao] |
| 19 |
Hb_000300_170 |
0.1293960101 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 20 |
Hb_001799_090 |
0.1297979006 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |