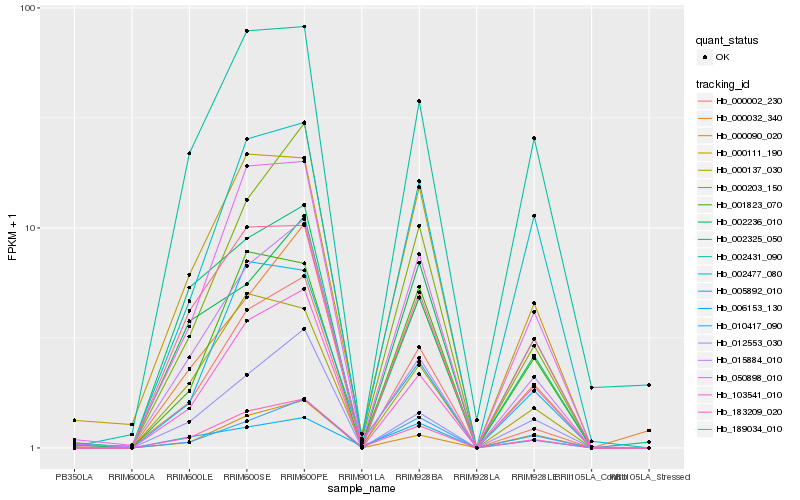
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183209_020 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 2 |
Hb_050898_010 |
0.093455872 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 3 |
Hb_103541_010 |
0.1012716699 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 4 |
Hb_189034_010 |
0.1144974149 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000002_230 |
0.1152985364 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 6 |
Hb_000137_030 |
0.1167851739 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Citrus sinensis] |
| 7 |
Hb_000203_150 |
0.1196653443 |
- |
- |
PREDICTED: uncharacterized protein LOC105116038 [Populus euphratica] |
| 8 |
Hb_015884_010 |
0.1240426438 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000111_190 |
0.1262193283 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 10 |
Hb_000032_340 |
0.1263069006 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 11 |
Hb_002431_090 |
0.1284733263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002477_080 |
0.1291619938 |
- |
- |
hypothetical protein RCOM_0653450 [Ricinus communis] |
| 13 |
Hb_005892_010 |
0.1301653144 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 14 |
Hb_000090_020 |
0.130240359 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 15 |
Hb_012553_030 |
0.1303513718 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 16 |
Hb_002325_050 |
0.1315550324 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 17 |
Hb_010417_090 |
0.1413195589 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 18 |
Hb_001823_070 |
0.1464957456 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 19 |
Hb_002236_010 |
0.1466678913 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |
| 20 |
Hb_006153_130 |
0.1467838275 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |