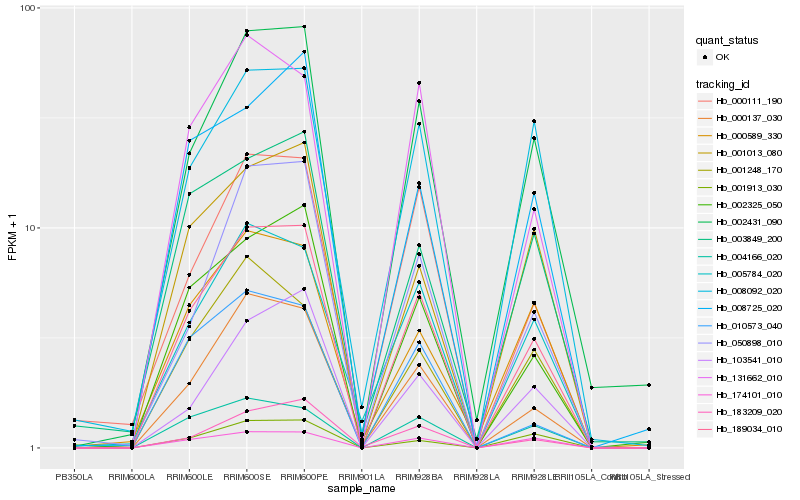
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_189034_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000137_030 |
0.078606461 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Citrus sinensis] |
| 3 |
Hb_002431_090 |
0.0880456846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002325_050 |
0.0933440617 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 5 |
Hb_005784_020 |
0.11416713 |
- |
- |
Protein MKS1, putative [Ricinus communis] |
| 6 |
Hb_183209_020 |
0.1144974149 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 7 |
Hb_050898_010 |
0.1164318747 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 8 |
Hb_131662_010 |
0.1181629836 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 9 |
Hb_004166_020 |
0.1204141816 |
- |
- |
- |
| 10 |
Hb_001013_080 |
0.1230479823 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 11 |
Hb_000111_190 |
0.1250580965 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 12 |
Hb_174101_010 |
0.1259021857 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 13 |
Hb_103541_010 |
0.1282428101 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 14 |
Hb_008725_020 |
0.1312653646 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 15 |
Hb_001913_030 |
0.1320255977 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 16 |
Hb_001248_170 |
0.1329412499 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 17 |
Hb_008092_020 |
0.1334471907 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_010573_040 |
0.1348262493 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 19 |
Hb_000589_330 |
0.1367720111 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_003849_200 |
0.1375085189 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |