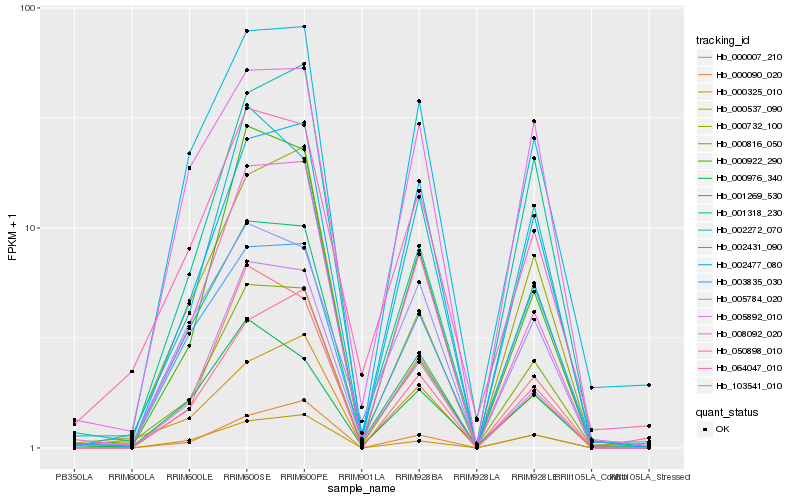
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_050 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 2 |
Hb_064047_010 |
0.0990018475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_050898_010 |
0.0994875498 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 4 |
Hb_002477_080 |
0.1065348474 |
- |
- |
hypothetical protein RCOM_0653450 [Ricinus communis] |
| 5 |
Hb_002431_090 |
0.1078741849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000325_010 |
0.117630791 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 7 |
Hb_002272_070 |
0.1196537007 |
- |
- |
PREDICTED: keratin, type I cytoskeletal 9 [Jatropha curcas] |
| 8 |
Hb_000732_100 |
0.1229303092 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 9 |
Hb_003835_030 |
0.123714498 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 10 |
Hb_103541_010 |
0.1238957897 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000007_210 |
0.1244004417 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 12 |
Hb_001269_530 |
0.125849057 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 13 |
Hb_000922_290 |
0.1263614103 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 14 |
Hb_000976_340 |
0.1267457402 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_008092_020 |
0.1295924316 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000537_090 |
0.1296187921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005892_010 |
0.1301310504 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 18 |
Hb_000090_020 |
0.1311246591 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 19 |
Hb_005784_020 |
0.132947837 |
- |
- |
Protein MKS1, putative [Ricinus communis] |
| 20 |
Hb_001318_230 |
0.1331336065 |
- |
- |
Boron transporter, putative [Ricinus communis] |