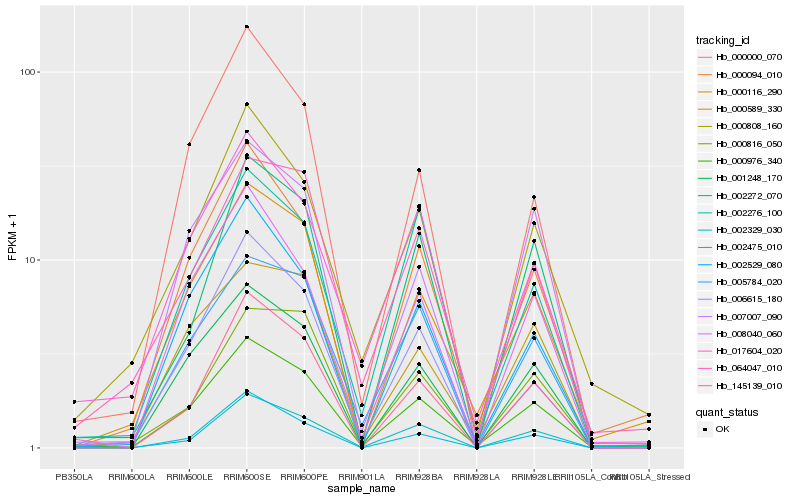
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000976_340 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000116_290 |
0.0907704276 |
- |
- |
PREDICTED: proton-coupled amino acid transporter 3 [Jatropha curcas] |
| 3 |
Hb_001248_170 |
0.093648546 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 4 |
Hb_000094_010 |
0.1037872762 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_005784_020 |
0.1074226064 |
- |
- |
Protein MKS1, putative [Ricinus communis] |
| 6 |
Hb_008040_060 |
0.1130851507 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 7 |
Hb_002329_030 |
0.1135239321 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000589_330 |
0.114003763 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_145139_010 |
0.1148260879 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 isoform X1 [Vitis vinifera] |
| 10 |
Hb_007007_090 |
0.1152580736 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 11 |
Hb_017604_020 |
0.1176672559 |
- |
- |
hypothetical protein JCGZ_22673 [Jatropha curcas] |
| 12 |
Hb_000808_160 |
0.1179766977 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 13 |
Hb_002529_080 |
0.1195950391 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 14 |
Hb_064047_010 |
0.1203099307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006615_180 |
0.1205584223 |
- |
- |
hypothetical protein B456_003G103400 [Gossypium raimondii] |
| 16 |
Hb_002272_070 |
0.1225137788 |
- |
- |
PREDICTED: keratin, type I cytoskeletal 9 [Jatropha curcas] |
| 17 |
Hb_000000_070 |
0.1253875522 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |
| 18 |
Hb_002475_010 |
0.1261639057 |
- |
- |
hypothetical protein JCGZ_22530 [Jatropha curcas] |
| 19 |
Hb_000816_050 |
0.1267457402 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 20 |
Hb_002276_100 |
0.1287449304 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89A2-like [Jatropha curcas] |