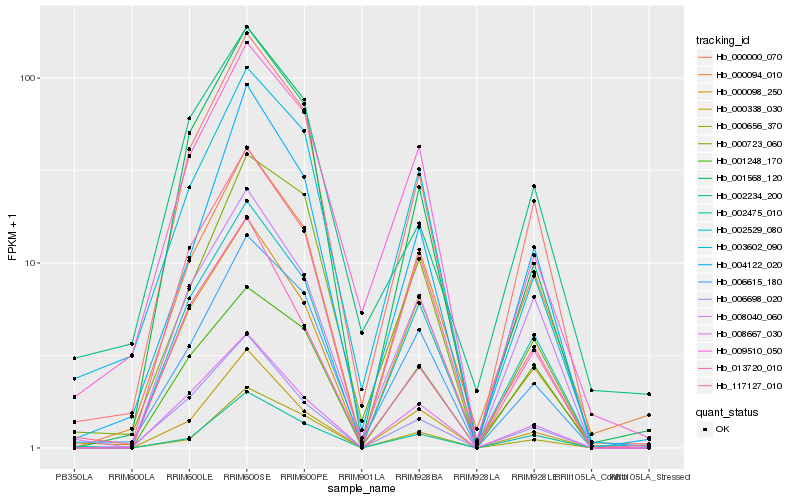
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_070 |
0.0 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |
| 2 |
Hb_006615_180 |
0.0759375036 |
- |
- |
hypothetical protein B456_003G103400 [Gossypium raimondii] |
| 3 |
Hb_002529_080 |
0.0771252028 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 4 |
Hb_001568_120 |
0.0948250804 |
- |
- |
hypothetical protein JCGZ_05918 [Jatropha curcas] |
| 5 |
Hb_117127_010 |
0.0950640771 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |
| 6 |
Hb_008667_030 |
0.0956357791 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 7 |
Hb_004122_020 |
0.0969429037 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 8 |
Hb_006698_020 |
0.0973626402 |
- |
- |
hypothetical protein JCGZ_04585 [Jatropha curcas] |
| 9 |
Hb_008040_060 |
0.1018783509 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 10 |
Hb_003602_090 |
0.1097927558 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 11 |
Hb_000094_010 |
0.1105565078 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_009510_050 |
0.1140778313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002475_010 |
0.1171425479 |
- |
- |
hypothetical protein JCGZ_22530 [Jatropha curcas] |
| 14 |
Hb_000098_250 |
0.1173012999 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 15 |
Hb_001248_170 |
0.1196534964 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 16 |
Hb_000723_060 |
0.120552692 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 17 |
Hb_000656_370 |
0.1210017894 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_013720_010 |
0.1210382905 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 19 |
Hb_000338_030 |
0.1212591584 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_21254 [Jatropha curcas] |
| 20 |
Hb_002234_200 |
0.1249666517 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |