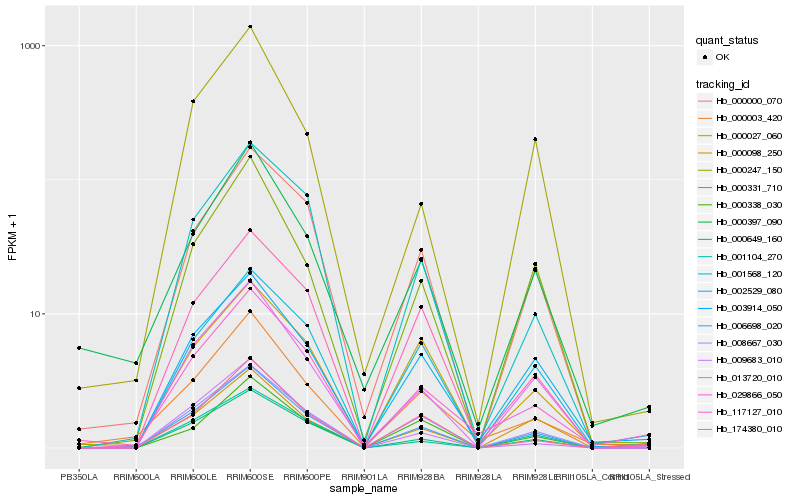
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006698_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_04585 [Jatropha curcas] |
| 2 |
Hb_013720_010 |
0.0818299305 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 3 |
Hb_008667_030 |
0.0839616832 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 4 |
Hb_000000_070 |
0.0973626402 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |
| 5 |
Hb_029866_050 |
0.1020261642 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 6 |
Hb_000331_710 |
0.1041742782 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_002529_080 |
0.1065543052 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 8 |
Hb_003914_050 |
0.1078855325 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 9 |
Hb_117127_010 |
0.1092326033 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |
| 10 |
Hb_001568_120 |
0.1092929837 |
- |
- |
hypothetical protein JCGZ_05918 [Jatropha curcas] |
| 11 |
Hb_000027_060 |
0.1134760105 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 12 |
Hb_001104_270 |
0.1135138211 |
- |
- |
hypothetical protein POPTR_0007s13980g [Populus trichocarpa] |
| 13 |
Hb_000003_420 |
0.1182549829 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_174380_010 |
0.1199957303 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_000649_160 |
0.1205380767 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000397_090 |
0.1247892693 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 17 |
Hb_009683_010 |
0.1255203039 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_000338_030 |
0.1258370918 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_21254 [Jatropha curcas] |
| 19 |
Hb_000247_150 |
0.1314206102 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 20 |
Hb_000098_250 |
0.131849708 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |