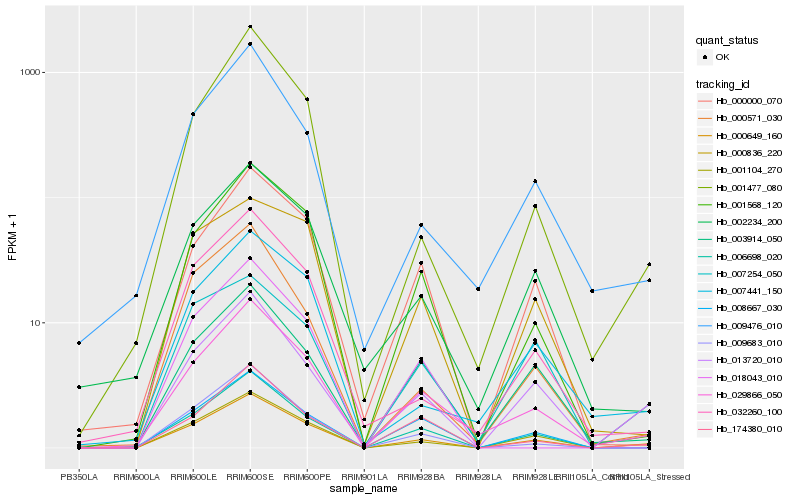
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_160 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 2 |
Hb_029866_050 |
0.1077585813 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 3 |
Hb_006698_020 |
0.1205380767 |
- |
- |
hypothetical protein JCGZ_04585 [Jatropha curcas] |
| 4 |
Hb_001104_270 |
0.1250881823 |
- |
- |
hypothetical protein POPTR_0007s13980g [Populus trichocarpa] |
| 5 |
Hb_001568_120 |
0.1324907969 |
- |
- |
hypothetical protein JCGZ_05918 [Jatropha curcas] |
| 6 |
Hb_013720_010 |
0.1409364341 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 7 |
Hb_007254_050 |
0.146936646 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 8 |
Hb_000000_070 |
0.1494067232 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |
| 9 |
Hb_174380_010 |
0.1496191343 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_007441_150 |
0.1510575316 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 11 |
Hb_002234_200 |
0.1527359455 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 12 |
Hb_001477_080 |
0.1529345777 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 13 |
Hb_009683_010 |
0.1535375187 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_009476_010 |
0.1567089017 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 15 |
Hb_008667_030 |
0.1567410596 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 16 |
Hb_003914_050 |
0.1568708348 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 17 |
Hb_032260_100 |
0.1599267188 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 18 |
Hb_000836_220 |
0.160488823 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000571_030 |
0.161107231 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 20 |
Hb_018043_010 |
0.1613234237 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |