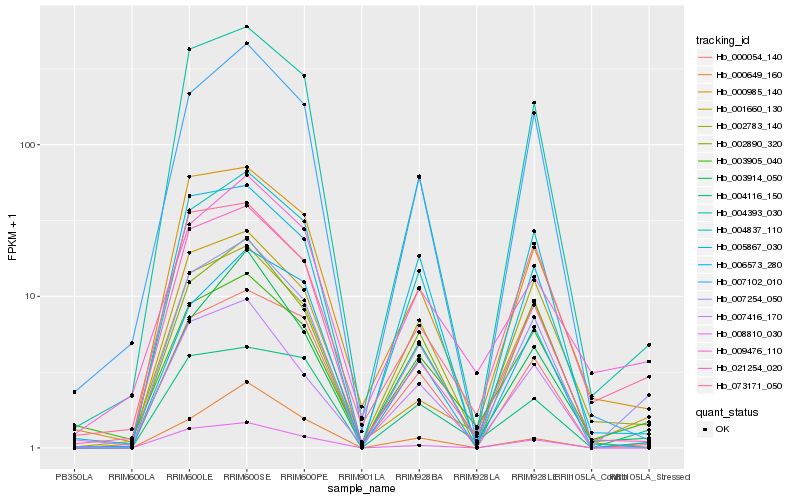
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007254_050 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 2 |
Hb_004393_030 |
0.1164736713 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 3 |
Hb_003905_040 |
0.1235954168 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 4 |
Hb_000054_140 |
0.125757264 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 5 |
Hb_004837_110 |
0.1269876405 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000985_140 |
0.1291532501 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 7 |
Hb_004116_150 |
0.1310325548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002890_320 |
0.1319045544 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 9 |
Hb_007102_010 |
0.1329884658 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 10 |
Hb_005867_030 |
0.1357025848 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 11 |
Hb_001660_130 |
0.1361718791 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR4 [Jatropha curcas] |
| 12 |
Hb_008810_030 |
0.1378760118 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_003914_050 |
0.1414872065 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 14 |
Hb_021254_020 |
0.142207954 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_073171_050 |
0.1426390037 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 16 |
Hb_006573_280 |
0.1454913004 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 17 |
Hb_009476_110 |
0.1456041218 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 18 |
Hb_007416_170 |
0.1467412251 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 19 |
Hb_000649_160 |
0.146936646 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 20 |
Hb_002783_140 |
0.1478168384 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |