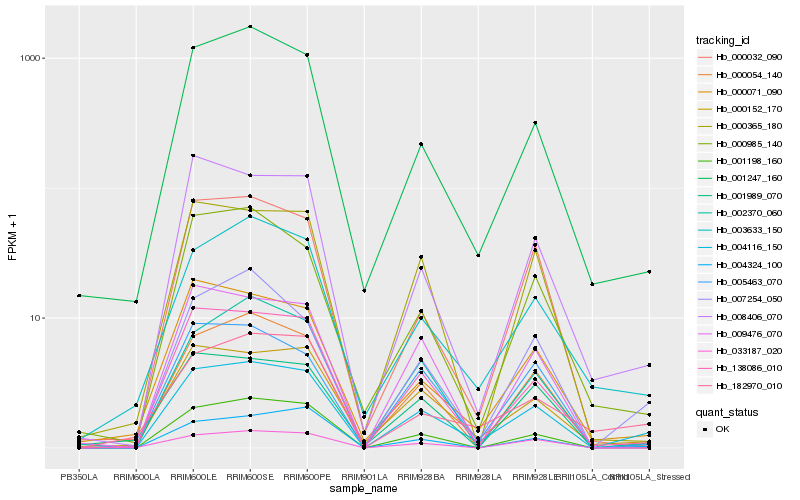
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004116_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_138086_010 |
0.1206660786 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 3 |
Hb_000152_170 |
0.1261176122 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 4 |
Hb_007254_050 |
0.1310325548 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 5 |
Hb_000054_140 |
0.1312983898 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 6 |
Hb_001247_160 |
0.14117196 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 7 |
Hb_008406_070 |
0.1438924494 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 8 |
Hb_001198_160 |
0.1472849594 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 9 |
Hb_003633_150 |
0.1474551298 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 10 |
Hb_004324_100 |
0.1498499222 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 11 |
Hb_005463_070 |
0.1506027376 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_002370_060 |
0.1520505753 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 13 |
Hb_001989_070 |
0.1534074508 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 14 |
Hb_000071_090 |
0.1542302937 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 15 |
Hb_000365_180 |
0.1546059552 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 16 |
Hb_000985_140 |
0.1569199515 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 17 |
Hb_182970_010 |
0.1572123938 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 18 |
Hb_033187_020 |
0.1586539458 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 19 |
Hb_009476_070 |
0.1593764885 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 20 |
Hb_000032_090 |
0.1601294901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |