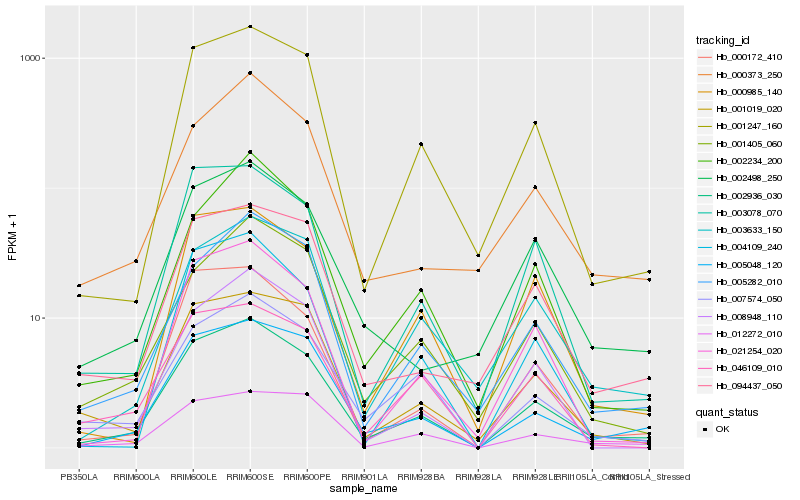
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002936_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005048_120 |
0.0924598488 |
- |
- |
- |
| 3 |
Hb_001247_160 |
0.1057014436 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 4 |
Hb_007574_050 |
0.1190066479 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 5 |
Hb_005282_010 |
0.1196847362 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 6 |
Hb_003078_070 |
0.1204632124 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g25290 [Jatropha curcas] |
| 7 |
Hb_012272_010 |
0.1256985726 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 8 |
Hb_001405_060 |
0.1259648344 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 9 |
Hb_000172_410 |
0.1260216459 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_021254_020 |
0.1265919394 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_001019_020 |
0.1273773305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_094437_050 |
0.1297625843 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003633_150 |
0.1298129993 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 14 |
Hb_000985_140 |
0.1323883468 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 15 |
Hb_002234_200 |
0.1325929345 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 16 |
Hb_000373_250 |
0.1327775006 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 17 |
Hb_008948_110 |
0.1346774891 |
- |
- |
PREDICTED: peroxidase 25 [Jatropha curcas] |
| 18 |
Hb_004109_240 |
0.1368312579 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 19 |
Hb_002498_250 |
0.137010095 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_046109_010 |
0.1385499869 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |