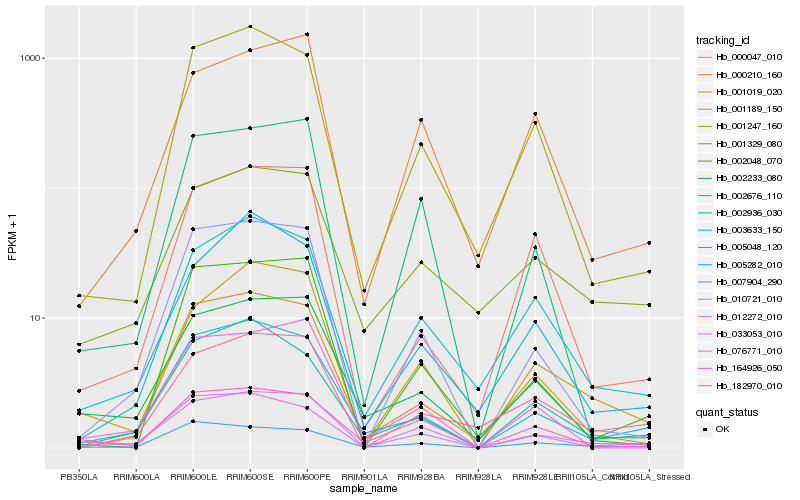
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012272_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 2 |
Hb_005048_120 |
0.1060847719 |
- |
- |
- |
| 3 |
Hb_010721_010 |
0.1196679647 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 4 |
Hb_007904_290 |
0.1204332755 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 5 |
Hb_001019_020 |
0.1214337903 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001189_150 |
0.1226401424 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 89-like [Jatropha curcas] |
| 7 |
Hb_002936_030 |
0.1256985726 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003633_150 |
0.1277711553 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 9 |
Hb_002233_080 |
0.1281180608 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 10 |
Hb_001247_160 |
0.1315856722 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 11 |
Hb_000210_160 |
0.1318966661 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 12 |
Hb_001329_080 |
0.1348534497 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 13 |
Hb_033053_010 |
0.1351734776 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 14 |
Hb_076771_010 |
0.1355593744 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 15 |
Hb_002676_110 |
0.1395010707 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 2-like [Gossypium raimondii] |
| 16 |
Hb_164926_050 |
0.1399613567 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 17 |
Hb_182970_010 |
0.1408458096 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 18 |
Hb_000047_010 |
0.1443015277 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 19 |
Hb_005282_010 |
0.1460710473 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 20 |
Hb_002048_070 |
0.1470076486 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |