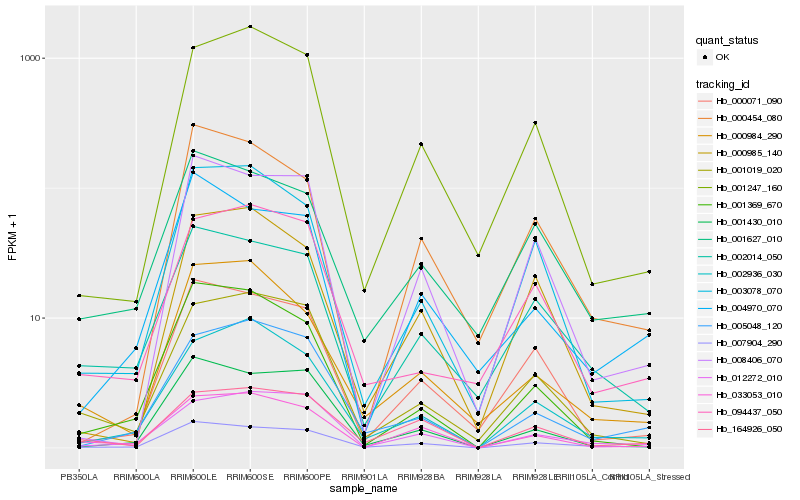
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007904_290 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 2 |
Hb_002014_050 |
0.1191761532 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 3 |
Hb_001430_010 |
0.1200883276 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_012272_010 |
0.1204332755 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 5 |
Hb_000071_090 |
0.122780796 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 6 |
Hb_001627_010 |
0.1258262881 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 7 |
Hb_004970_070 |
0.1267490689 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 8 |
Hb_008406_070 |
0.1280044659 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 9 |
Hb_033053_010 |
0.1334998687 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 10 |
Hb_005048_120 |
0.1354592131 |
- |
- |
- |
| 11 |
Hb_003078_070 |
0.1401743744 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g25290 [Jatropha curcas] |
| 12 |
Hb_002936_030 |
0.1408133224 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 13 |
Hb_164926_050 |
0.1417813269 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 14 |
Hb_000454_080 |
0.142956964 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 15 |
Hb_001019_020 |
0.1431593606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001369_670 |
0.1471580829 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 17 |
Hb_000985_140 |
0.1512342641 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000984_290 |
0.1518081964 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001247_160 |
0.1519202697 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 20 |
Hb_094437_050 |
0.1575094353 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |