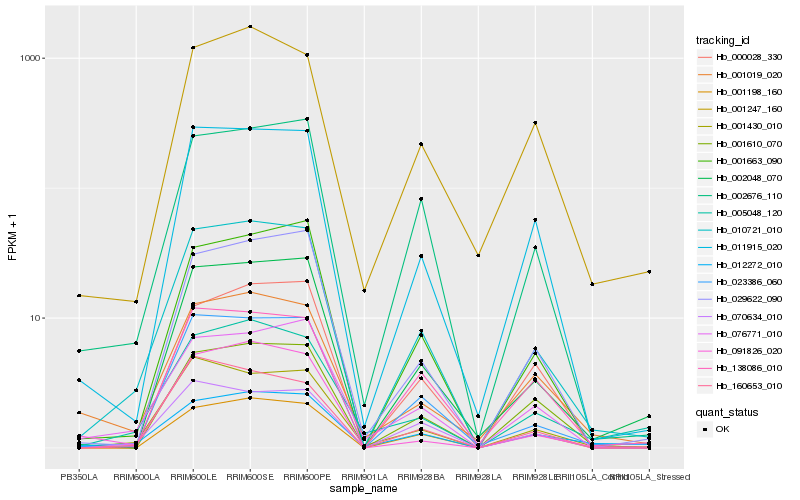
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010721_010 |
0.0 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 2 |
Hb_023386_060 |
0.0857534622 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 3 |
Hb_002048_070 |
0.1023332954 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |
| 4 |
Hb_001430_010 |
0.1023778829 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002676_110 |
0.1039552685 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 2-like [Gossypium raimondii] |
| 6 |
Hb_011915_020 |
0.1047877683 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 6, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_001663_090 |
0.1078532634 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 8 |
Hb_029622_090 |
0.1127994378 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2-like [Jatropha curcas] |
| 9 |
Hb_076771_010 |
0.1172029385 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 10 |
Hb_012272_010 |
0.1196679647 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_070634_010 |
0.121320403 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 12 |
Hb_001610_070 |
0.1258089992 |
- |
- |
hypothetical protein MIMGU_mgv1a022867mg [Erythranthe guttata] |
| 13 |
Hb_005048_120 |
0.1259763662 |
- |
- |
- |
| 14 |
Hb_001198_160 |
0.1264053764 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 15 |
Hb_160653_010 |
0.1278903229 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 16 |
Hb_001019_020 |
0.1328352728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001247_160 |
0.1333049839 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 18 |
Hb_091826_020 |
0.1335526824 |
- |
- |
hypothetical protein JCGZ_04975 [Jatropha curcas] |
| 19 |
Hb_138086_010 |
0.1341634508 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 20 |
Hb_000028_330 |
0.135303722 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |