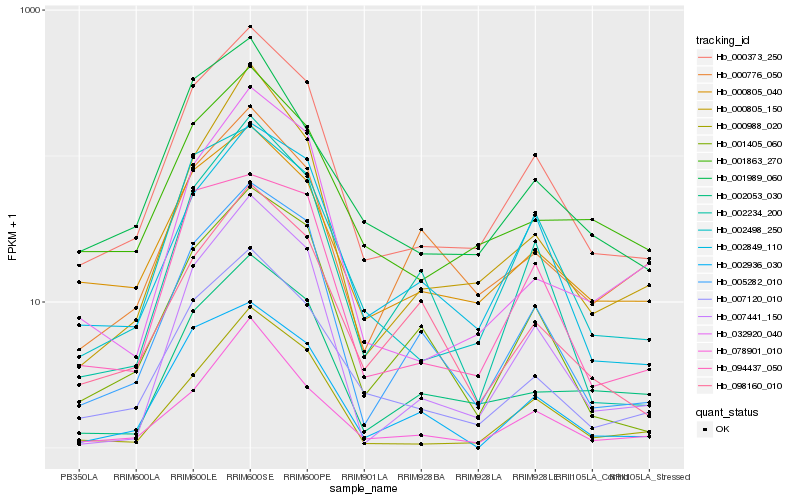
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000373_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 2 |
Hb_007120_010 |
0.0746642997 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |
| 3 |
Hb_005282_010 |
0.1019470436 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 4 |
Hb_002498_250 |
0.1043873114 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_000988_020 |
0.1095666586 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002849_110 |
0.11055468 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 7 |
Hb_001405_060 |
0.1158117017 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 8 |
Hb_000805_040 |
0.1164615268 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 9 |
Hb_000805_150 |
0.1164885509 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_078901_010 |
0.1181202021 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 11 |
Hb_001863_270 |
0.1220495286 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 12 |
Hb_002234_200 |
0.1250975028 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 13 |
Hb_001989_060 |
0.1269322675 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002053_030 |
0.1274448185 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_094437_050 |
0.1286050321 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007441_150 |
0.1286160354 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 17 |
Hb_002936_030 |
0.1327775006 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 18 |
Hb_032920_040 |
0.1341180566 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s30260g [Populus trichocarpa] |
| 19 |
Hb_098160_010 |
0.1380395158 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000776_050 |
0.1434901846 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |