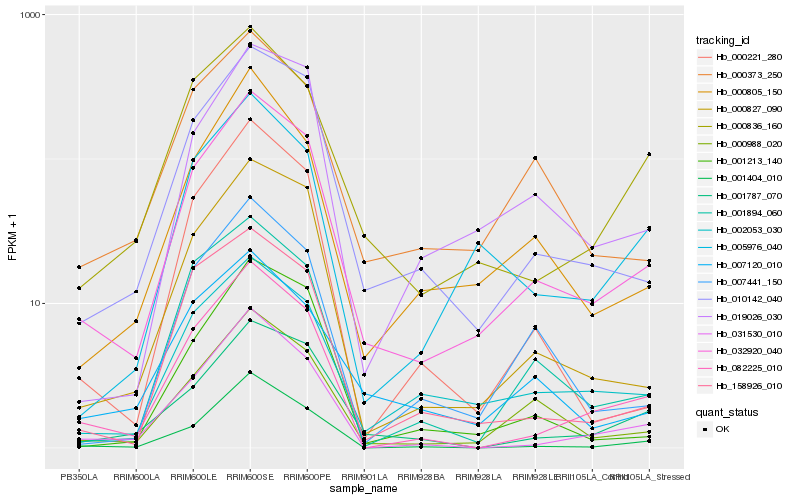
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032920_040 |
0.0 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s30260g [Populus trichocarpa] |
| 2 |
Hb_010142_040 |
0.0945005428 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 3 |
Hb_000827_090 |
0.1086123648 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 4 |
Hb_000988_020 |
0.1116589403 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 5 |
Hb_082225_010 |
0.113882669 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 6 |
Hb_000805_150 |
0.121426034 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_002053_030 |
0.1265052602 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_031530_010 |
0.1288739917 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 9 |
Hb_001787_070 |
0.1319013227 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Camelina sativa] |
| 10 |
Hb_000836_160 |
0.1319665689 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 11 |
Hb_000373_250 |
0.1341180566 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 12 |
Hb_019026_030 |
0.1375269002 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 13 |
Hb_158926_010 |
0.1383831411 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 14 |
Hb_001894_060 |
0.1398339313 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 15 |
Hb_001404_010 |
0.141390336 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 16 |
Hb_005976_040 |
0.1414757427 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 17 |
Hb_007441_150 |
0.1421636485 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 18 |
Hb_001213_140 |
0.1433531703 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 19 |
Hb_000221_280 |
0.1467497351 |
- |
- |
PREDICTED: protein EXORDIUM-like [Populus euphratica] |
| 20 |
Hb_007120_010 |
0.1474145138 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |