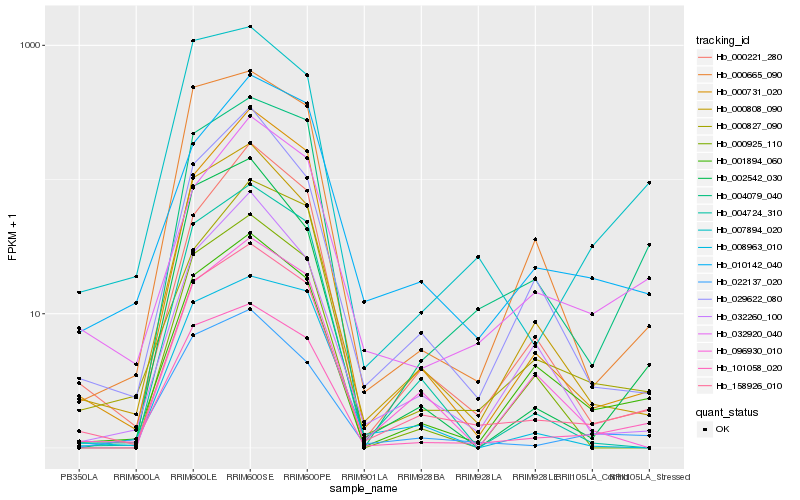
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158926_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 2 |
Hb_101058_020 |
0.0738051761 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 3 |
Hb_001894_060 |
0.1077940565 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 4 |
Hb_000808_090 |
0.1105281567 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 5 |
Hb_022137_020 |
0.1107182419 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 6 |
Hb_004079_040 |
0.1119950336 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 7 |
Hb_000665_090 |
0.1227599787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000731_020 |
0.1243287024 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 9 |
Hb_000221_280 |
0.1253821112 |
- |
- |
PREDICTED: protein EXORDIUM-like [Populus euphratica] |
| 10 |
Hb_007894_020 |
0.1260429929 |
- |
- |
protein translation factor SUI1 homolog 2 [Jatropha curcas] |
| 11 |
Hb_004724_310 |
0.1273967732 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 12 |
Hb_029622_080 |
0.1290744177 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 13 |
Hb_010142_040 |
0.1339593137 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 14 |
Hb_000827_090 |
0.1360731014 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 15 |
Hb_032920_040 |
0.1383831411 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s30260g [Populus trichocarpa] |
| 16 |
Hb_008963_010 |
0.1412549019 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_096930_010 |
0.1449653044 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 18 |
Hb_032260_100 |
0.1454979495 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 19 |
Hb_002542_030 |
0.1460567445 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 20 |
Hb_000925_110 |
0.1469673622 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |