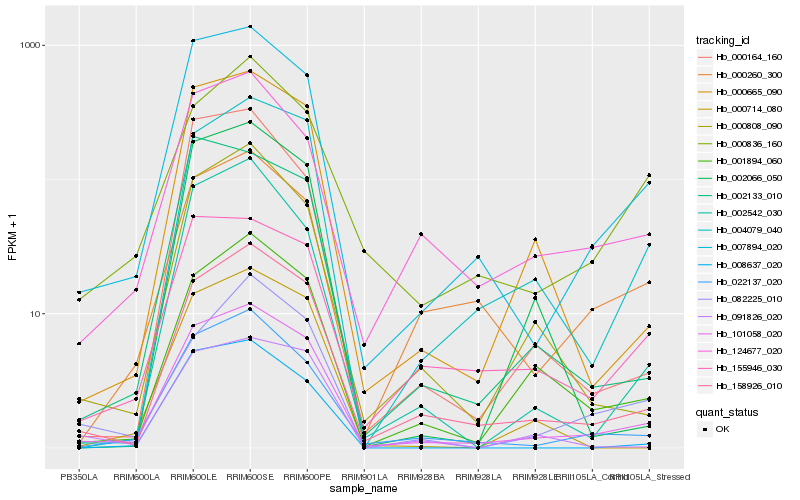
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101058_020 |
0.0 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 2 |
Hb_158926_010 |
0.0738051761 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 3 |
Hb_007894_020 |
0.0743176371 |
- |
- |
protein translation factor SUI1 homolog 2 [Jatropha curcas] |
| 4 |
Hb_022137_020 |
0.0961883336 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 5 |
Hb_000665_090 |
0.1123203412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001894_060 |
0.1177464332 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 7 |
Hb_004079_040 |
0.12267466 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 8 |
Hb_000808_090 |
0.1253975535 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 9 |
Hb_002542_030 |
0.1358786756 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 10 |
Hb_124677_020 |
0.1390949709 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 11 |
Hb_155946_030 |
0.1431709624 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 12 |
Hb_000714_080 |
0.1458505711 |
transcription factor |
TF Family: Trihelix |
GT-2 factor [Glycine max] |
| 13 |
Hb_091826_020 |
0.1491042547 |
- |
- |
hypothetical protein JCGZ_04975 [Jatropha curcas] |
| 14 |
Hb_002066_050 |
0.1520192465 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 15 |
Hb_002133_010 |
0.152244866 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 16 |
Hb_082225_010 |
0.152798749 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 17 |
Hb_000164_160 |
0.1532139565 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_008637_020 |
0.1534358646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000836_160 |
0.1552100927 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 20 |
Hb_000260_300 |
0.1552735025 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |