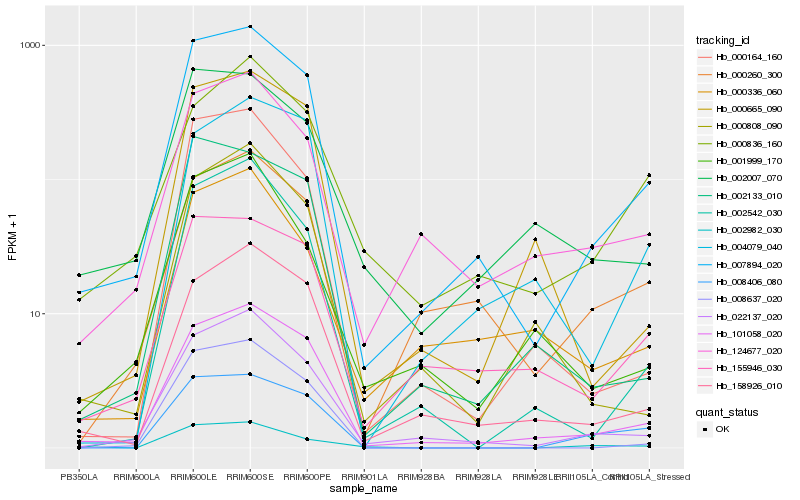
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_020 |
0.0 |
- |
- |
protein translation factor SUI1 homolog 2 [Jatropha curcas] |
| 2 |
Hb_101058_020 |
0.0743176371 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 3 |
Hb_022137_020 |
0.0944728312 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 4 |
Hb_158926_010 |
0.1260429929 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 5 |
Hb_002542_030 |
0.1370606644 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 6 |
Hb_155946_030 |
0.1384806759 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 7 |
Hb_124677_020 |
0.1390940199 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 8 |
Hb_008637_020 |
0.1417747274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000260_300 |
0.1454782051 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 10 |
Hb_000164_160 |
0.1458334221 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002133_010 |
0.1482581532 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 12 |
Hb_000665_090 |
0.1522907869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008406_080 |
0.1533312187 |
- |
- |
Nonspecific lipid-transfer protein 3 precursor, putative [Ricinus communis] |
| 14 |
Hb_002007_070 |
0.1548501384 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 15 |
Hb_002982_030 |
0.160209785 |
- |
- |
Chalcone synthase, putative [Ricinus communis] |
| 16 |
Hb_000336_060 |
0.1603192211 |
- |
- |
hypothetical protein JCGZ_26632 [Jatropha curcas] |
| 17 |
Hb_001999_170 |
0.1622912931 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 18 |
Hb_000808_090 |
0.1626208011 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 19 |
Hb_004079_040 |
0.1646701584 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 20 |
Hb_000836_160 |
0.1656210385 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |