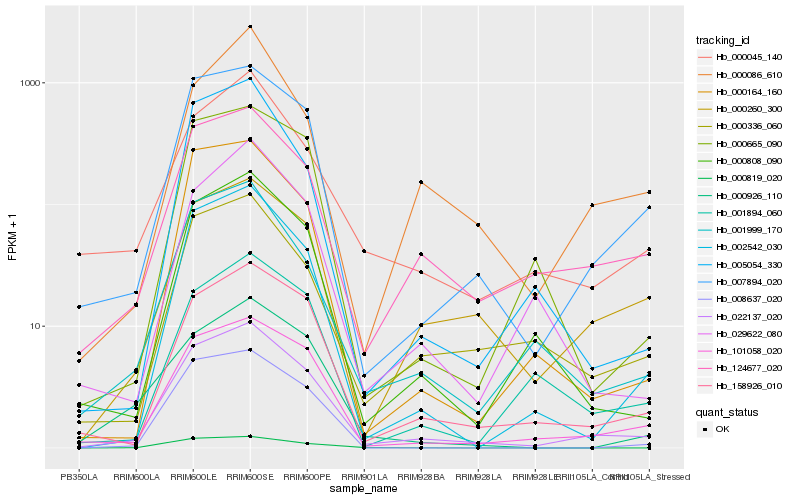
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022137_020 |
0.0 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 2 |
Hb_007894_020 |
0.0944728312 |
- |
- |
protein translation factor SUI1 homolog 2 [Jatropha curcas] |
| 3 |
Hb_101058_020 |
0.0961883336 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 4 |
Hb_158926_010 |
0.1107182419 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 5 |
Hb_002542_030 |
0.1165396525 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 6 |
Hb_124677_020 |
0.1184736167 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 7 |
Hb_001999_170 |
0.1225052329 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 8 |
Hb_000808_090 |
0.123622367 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 9 |
Hb_000164_160 |
0.1355515047 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000336_060 |
0.1378845324 |
- |
- |
hypothetical protein JCGZ_26632 [Jatropha curcas] |
| 11 |
Hb_005054_330 |
0.1447969469 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 12 |
Hb_000260_300 |
0.1459230243 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 13 |
Hb_000045_140 |
0.1459932299 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 14 |
Hb_008637_020 |
0.1468623609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000819_020 |
0.1508412737 |
- |
- |
hypothetical protein PRUPE_ppa020290mg, partial [Prunus persica] |
| 16 |
Hb_000665_090 |
0.1518676473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001894_060 |
0.1548356846 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 18 |
Hb_000926_110 |
0.1561170579 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 19 |
Hb_029622_080 |
0.1562747587 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 20 |
Hb_000086_610 |
0.1600436349 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |