| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
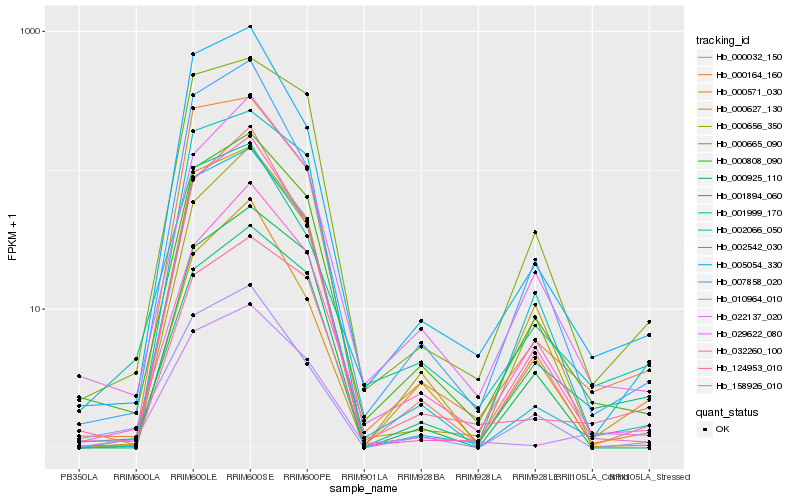
Hb_000808_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 2 |
Hb_029622_080 |
0.0739737786 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 3 |
Hb_000665_090 |
0.0946382509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_032260_100 |
0.0958104238 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 5 |
Hb_000656_350 |
0.0987777544 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000627_130 |
0.099267777 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 7 |
Hb_124953_010 |
0.0993865414 |
- |
- |
hypothetical protein RCOM_1314600 [Ricinus communis] |
| 8 |
Hb_000925_110 |
0.1056289999 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 9 |
Hb_002066_050 |
0.1086557638 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 10 |
Hb_158926_010 |
0.1105281567 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 11 |
Hb_010964_010 |
0.111942559 |
- |
- |
- |
| 12 |
Hb_001894_060 |
0.1131381982 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 13 |
Hb_007858_020 |
0.1133777235 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 14 |
Hb_002542_030 |
0.1146059999 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 15 |
Hb_005054_330 |
0.1153351201 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 16 |
Hb_001999_170 |
0.1171627594 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 17 |
Hb_000164_160 |
0.1191550586 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000032_150 |
0.1210286075 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1691430 [Ricinus communis] |
| 19 |
Hb_000571_030 |
0.1231830544 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 20 |
Hb_022137_020 |
0.123622367 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |