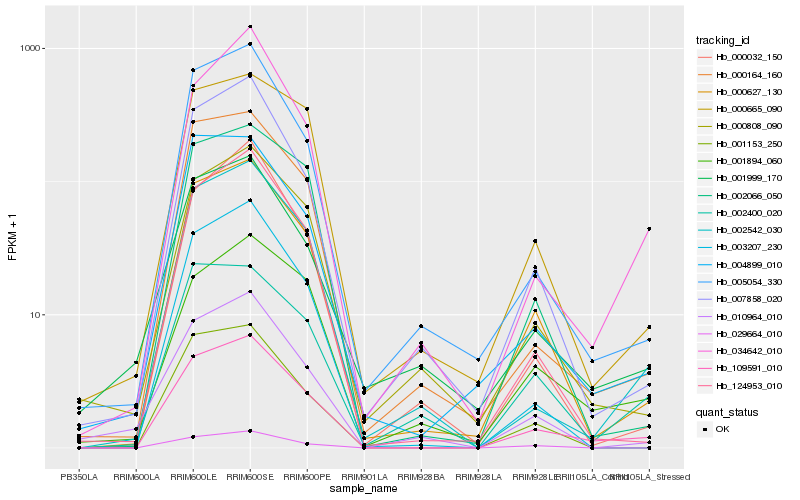
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_109591_010 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 2 |
Hb_000627_130 |
0.0903784368 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 3 |
Hb_002542_030 |
0.1274502949 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 4 |
Hb_005054_330 |
0.1278159527 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 5 |
Hb_007858_020 |
0.1282869142 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 6 |
Hb_000164_160 |
0.1285887333 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_124953_010 |
0.1299209898 |
- |
- |
hypothetical protein RCOM_1314600 [Ricinus communis] |
| 8 |
Hb_029664_010 |
0.1328081662 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_001153_250 |
0.1329392505 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 10 |
Hb_000808_090 |
0.1354150562 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 11 |
Hb_001894_060 |
0.1368899591 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 12 |
Hb_001999_170 |
0.1384447238 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 13 |
Hb_003207_230 |
0.1397063594 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 14 |
Hb_004899_010 |
0.1413407526 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 15 |
Hb_002400_020 |
0.1414317255 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 16 |
Hb_002066_050 |
0.1414609533 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 17 |
Hb_034642_010 |
0.1439854261 |
- |
- |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 18 |
Hb_000665_090 |
0.1487275643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000032_150 |
0.152084149 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1691430 [Ricinus communis] |
| 20 |
Hb_010964_010 |
0.153215522 |
- |
- |
- |