| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
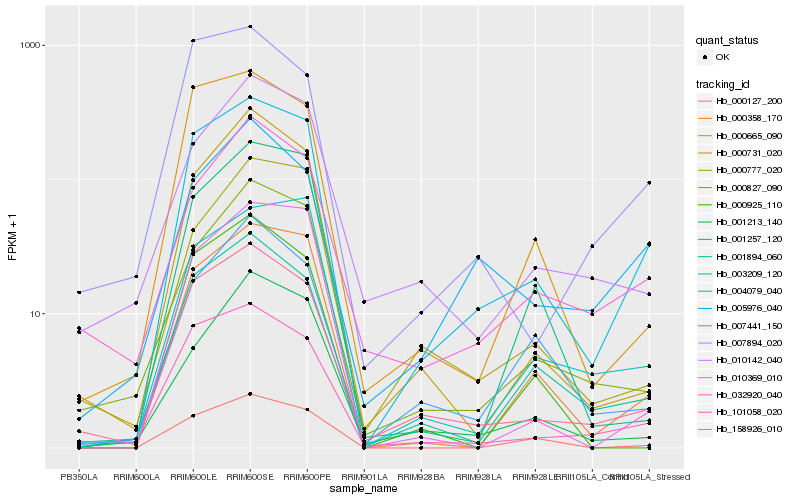
Hb_004079_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 2 |
Hb_000358_170 |
0.11155328 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 3 |
Hb_158926_010 |
0.1119950336 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 4 |
Hb_101058_020 |
0.12267466 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 5 |
Hb_001894_060 |
0.1280485352 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 6 |
Hb_000127_200 |
0.1367809455 |
- |
- |
PREDICTED: uncharacterized protein LOC100255537 [Vitis vinifera] |
| 7 |
Hb_000665_090 |
0.1441382137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003209_120 |
0.1464580445 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 9 |
Hb_000827_090 |
0.150396684 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 10 |
Hb_032920_040 |
0.1579696873 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s30260g [Populus trichocarpa] |
| 11 |
Hb_010369_010 |
0.1603205846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001257_120 |
0.1624965506 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 13 |
Hb_007894_020 |
0.1646701584 |
- |
- |
protein translation factor SUI1 homolog 2 [Jatropha curcas] |
| 14 |
Hb_000731_020 |
0.1649717245 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 15 |
Hb_000777_020 |
0.1658260701 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 16 |
Hb_007441_150 |
0.1659437331 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 17 |
Hb_001213_140 |
0.1659958089 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 18 |
Hb_000925_110 |
0.1669253459 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 19 |
Hb_010142_040 |
0.1677735953 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 20 |
Hb_005976_040 |
0.1677779094 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |