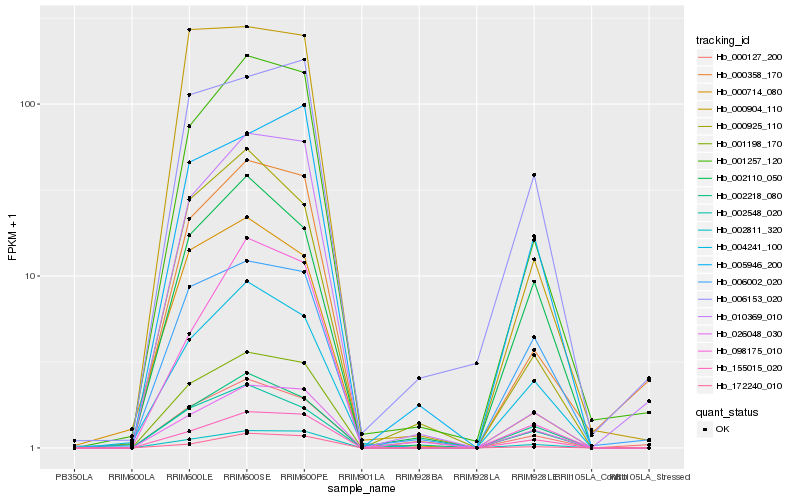
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001198_170 |
0.0 |
- |
- |
- |
| 2 |
Hb_001257_120 |
0.06846729 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 3 |
Hb_002811_320 |
0.0694726197 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 4 |
Hb_155015_020 |
0.0832575914 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 5 |
Hb_000127_200 |
0.0989150351 |
- |
- |
PREDICTED: uncharacterized protein LOC100255537 [Vitis vinifera] |
| 6 |
Hb_000358_170 |
0.1018851494 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 7 |
Hb_002548_020 |
0.1131307552 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004241_100 |
0.1145830036 |
- |
- |
PREDICTED: transcription factor bHLH123-like isoform X1 [Populus euphratica] |
| 9 |
Hb_172240_010 |
0.1168111528 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |
| 10 |
Hb_000925_110 |
0.12093942 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 11 |
Hb_005946_200 |
0.1298493376 |
- |
- |
hypothetical protein POPTR_0006s16210g [Populus trichocarpa] |
| 12 |
Hb_000714_080 |
0.1305055307 |
transcription factor |
TF Family: Trihelix |
GT-2 factor [Glycine max] |
| 13 |
Hb_000904_110 |
0.131123693 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Eucalyptus grandis] |
| 14 |
Hb_002218_080 |
0.1319327542 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 15 |
Hb_006153_020 |
0.1340437855 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 16 |
Hb_006002_020 |
0.1370256186 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 17 |
Hb_010369_010 |
0.1383884614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_026048_030 |
0.1394701603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_098175_010 |
0.1433493998 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 20 |
Hb_002110_050 |
0.1435000903 |
- |
- |
conserved hypothetical protein [Ricinus communis] |