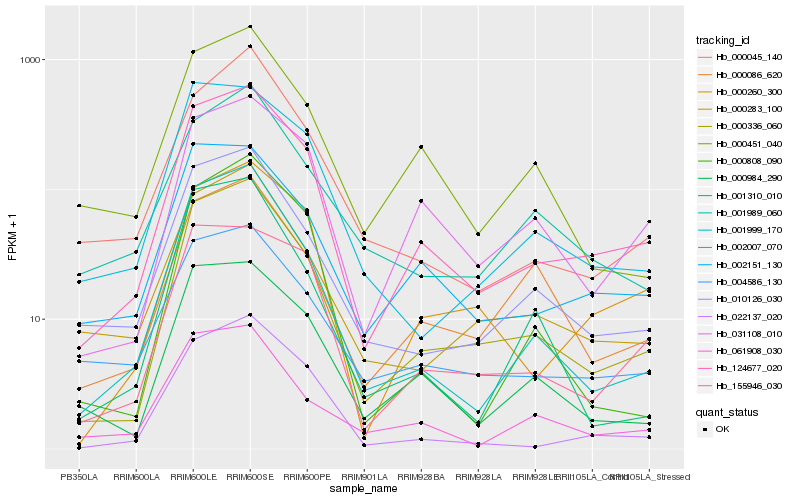
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000336_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_26632 [Jatropha curcas] |
| 2 |
Hb_124677_020 |
0.084028484 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 3 |
Hb_001999_170 |
0.1051330739 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 4 |
Hb_010126_030 |
0.1146337481 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000283_100 |
0.123811639 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000984_290 |
0.1335508457 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_022137_020 |
0.1378845324 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 8 |
Hb_000086_620 |
0.1383195421 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001310_010 |
0.1429384913 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 10 |
Hb_000260_300 |
0.1439901174 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 11 |
Hb_031108_010 |
0.1447223008 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 12 |
Hb_000451_040 |
0.1481030978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000808_090 |
0.1486840944 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 14 |
Hb_002151_130 |
0.1519724256 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 15 |
Hb_000045_140 |
0.152052126 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 16 |
Hb_002007_070 |
0.1522962041 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 17 |
Hb_061908_030 |
0.1526576736 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 18 |
Hb_001989_060 |
0.1539108324 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004586_130 |
0.1546301655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_155946_030 |
0.1556895388 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |