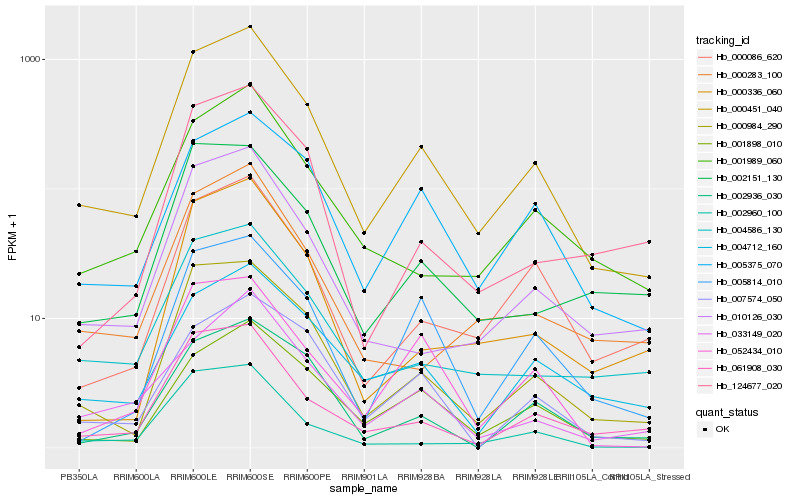
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000451_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000984_290 |
0.0997847444 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_061908_030 |
0.1260068387 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 4 |
Hb_010126_030 |
0.1286502247 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001989_060 |
0.1293352652 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004586_130 |
0.1298977095 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001898_010 |
0.1309664117 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 8 |
Hb_005375_070 |
0.1313427444 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 9 |
Hb_033149_020 |
0.1324675127 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 10 |
Hb_000086_620 |
0.1357083038 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004712_160 |
0.1396535533 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 12 |
Hb_000283_100 |
0.143270404 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_007574_050 |
0.1449553101 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 14 |
Hb_002151_130 |
0.1458844066 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 15 |
Hb_002936_030 |
0.1460997791 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002960_100 |
0.1467849461 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 17 |
Hb_000336_060 |
0.1481030978 |
- |
- |
hypothetical protein JCGZ_26632 [Jatropha curcas] |
| 18 |
Hb_052434_010 |
0.1485760837 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |
| 19 |
Hb_124677_020 |
0.1503093388 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 20 |
Hb_005814_010 |
0.1514287234 |
- |
- |
ATP binding protein, putative [Ricinus communis] |