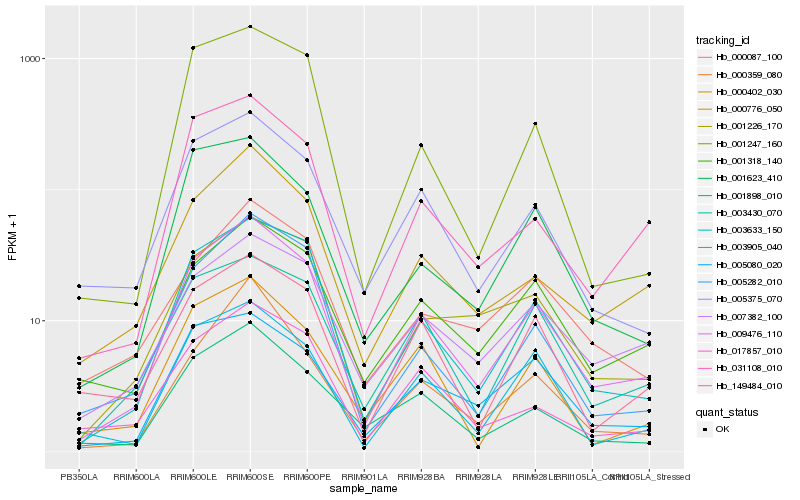
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009476_110 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 2 |
Hb_003633_150 |
0.0790304613 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 3 |
Hb_000776_050 |
0.1095806892 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 4 |
Hb_031108_010 |
0.1165755642 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 5 |
Hb_001898_010 |
0.1188165829 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 6 |
Hb_005375_070 |
0.1212389458 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 7 |
Hb_003905_040 |
0.1223986896 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 8 |
Hb_001318_140 |
0.1225110069 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 9 |
Hb_005282_010 |
0.124288749 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 10 |
Hb_001226_170 |
0.1262331234 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 11 |
Hb_001247_160 |
0.1271287902 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 12 |
Hb_003430_070 |
0.1275154635 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 13 |
Hb_007382_100 |
0.1304827888 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_005080_020 |
0.1305361108 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 15 |
Hb_001623_410 |
0.1305494271 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_149484_010 |
0.1311383404 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor-like [Eucalyptus grandis] |
| 17 |
Hb_017857_010 |
0.1313062235 |
- |
- |
- |
| 18 |
Hb_000359_080 |
0.1331387747 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 19 |
Hb_000402_030 |
0.1333568843 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 20 |
Hb_000087_100 |
0.1369449095 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |