| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
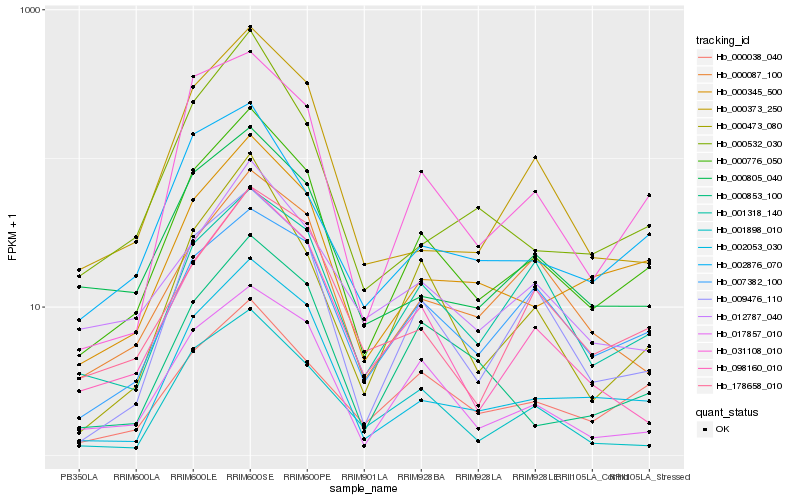
Hb_000776_050 |
0.0 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 2 |
Hb_002053_030 |
0.1025170986 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000345_500 |
0.1085363954 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 4 |
Hb_009476_110 |
0.1095806892 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 5 |
Hb_017857_010 |
0.1147589215 |
- |
- |
- |
| 6 |
Hb_031108_010 |
0.1166835494 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 7 |
Hb_000473_080 |
0.12568944 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 8 |
Hb_098160_010 |
0.1292322905 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000805_040 |
0.1298375727 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 10 |
Hb_007382_100 |
0.1317891949 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000038_040 |
0.1338201099 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 12 |
Hb_001898_010 |
0.1364316411 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 13 |
Hb_001318_140 |
0.1383769955 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 14 |
Hb_002876_070 |
0.1389316994 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 15 |
Hb_012787_040 |
0.1412170036 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000853_100 |
0.1414128216 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 17 |
Hb_178658_010 |
0.1431662314 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 18 |
Hb_000087_100 |
0.1433562424 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 19 |
Hb_000373_250 |
0.1434901846 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 20 |
Hb_000532_030 |
0.1440514545 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |