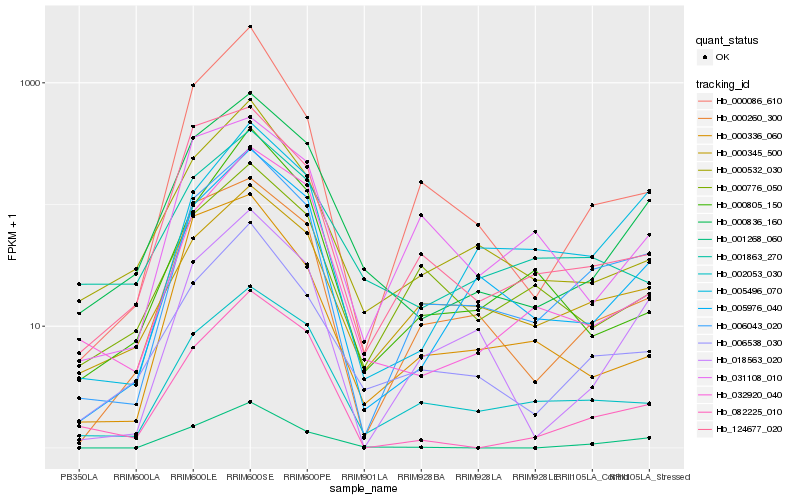
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006043_020 |
0.0 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |
| 2 |
Hb_002053_030 |
0.1180431737 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_005976_040 |
0.1198464468 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 4 |
Hb_000260_300 |
0.1226265999 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 5 |
Hb_000345_500 |
0.1319122009 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 6 |
Hb_018563_020 |
0.1347900198 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 6 [Hevea brasiliensis] |
| 7 |
Hb_006538_030 |
0.1387759141 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 8 |
Hb_000086_610 |
0.1521584251 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 9 |
Hb_005496_070 |
0.1543362788 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 10 |
Hb_124677_020 |
0.1612561567 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 11 |
Hb_000836_160 |
0.1623188092 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 12 |
Hb_000532_030 |
0.1662811061 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 13 |
Hb_000776_050 |
0.1670721787 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 14 |
Hb_032920_040 |
0.178379681 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s30260g [Populus trichocarpa] |
| 15 |
Hb_031108_010 |
0.1798808253 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 16 |
Hb_001268_060 |
0.1816865561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_082225_010 |
0.1828915477 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 18 |
Hb_000805_150 |
0.1847118101 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000336_060 |
0.1875975172 |
- |
- |
hypothetical protein JCGZ_26632 [Jatropha curcas] |
| 20 |
Hb_001863_270 |
0.1937402755 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |