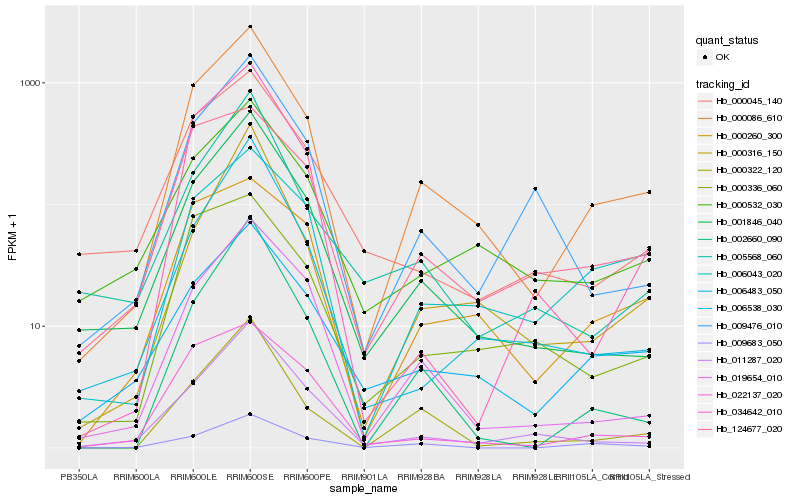
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_610 |
0.0 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 2 |
Hb_001846_040 |
0.1297043595 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_006538_030 |
0.1360424459 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 4 |
Hb_019654_010 |
0.1377984979 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 5 |
Hb_000322_120 |
0.1383469292 |
- |
- |
PREDICTED: uncharacterized protein LOC105637900 [Jatropha curcas] |
| 6 |
Hb_011287_020 |
0.1422976118 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_009683_050 |
0.1469796587 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 8 |
Hb_000532_030 |
0.1488047588 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 9 |
Hb_002660_090 |
0.149352832 |
- |
- |
hypothetical protein JCGZ_16500 [Jatropha curcas] |
| 10 |
Hb_006043_020 |
0.1521584251 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |
| 11 |
Hb_000316_150 |
0.1552794553 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 12 |
Hb_006483_050 |
0.1566363079 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 13 |
Hb_124677_020 |
0.1591349454 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 14 |
Hb_022137_020 |
0.1600436349 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 15 |
Hb_000260_300 |
0.161101247 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 16 |
Hb_000336_060 |
0.1622794292 |
- |
- |
hypothetical protein JCGZ_26632 [Jatropha curcas] |
| 17 |
Hb_034642_010 |
0.1631940607 |
- |
- |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 18 |
Hb_009476_010 |
0.1647148627 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 19 |
Hb_000045_140 |
0.1652521275 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 20 |
Hb_005568_060 |
0.1662631524 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |