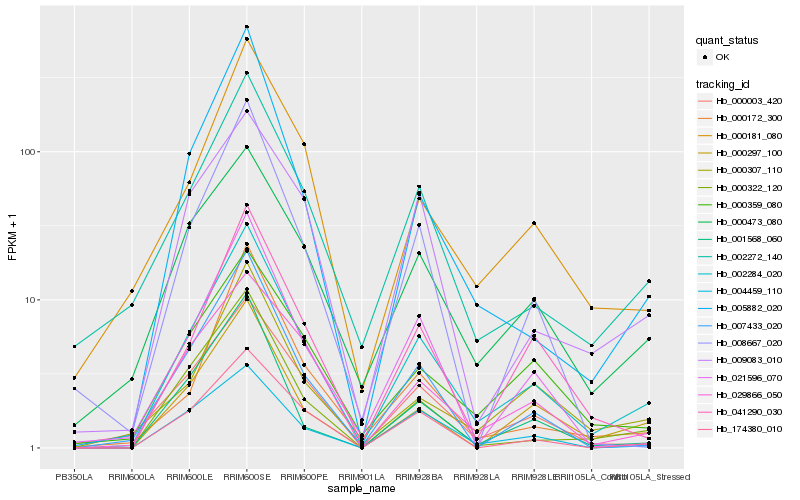
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_020 |
0.0 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 2 |
Hb_002272_140 |
0.122010899 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 3 |
Hb_000473_080 |
0.1272529998 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 4 |
Hb_000322_120 |
0.1275318821 |
- |
- |
PREDICTED: uncharacterized protein LOC105637900 [Jatropha curcas] |
| 5 |
Hb_174380_010 |
0.1279687441 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_000003_420 |
0.1306591328 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_021596_070 |
0.1321336189 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_000297_100 |
0.1324709525 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 9 |
Hb_000181_080 |
0.1325162629 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 10 |
Hb_007433_020 |
0.1325576038 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 11 |
Hb_041290_030 |
0.1332934016 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 12 |
Hb_029866_050 |
0.1347388755 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 13 |
Hb_000307_110 |
0.1349797571 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 14 |
Hb_000359_080 |
0.1416032652 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 15 |
Hb_009083_010 |
0.1442417683 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 16 |
Hb_000172_300 |
0.1451938156 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 17 |
Hb_001568_060 |
0.1454811402 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 18 |
Hb_008667_020 |
0.1460951969 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 19 |
Hb_004459_110 |
0.1472877332 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 20 |
Hb_005882_020 |
0.1479746056 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |