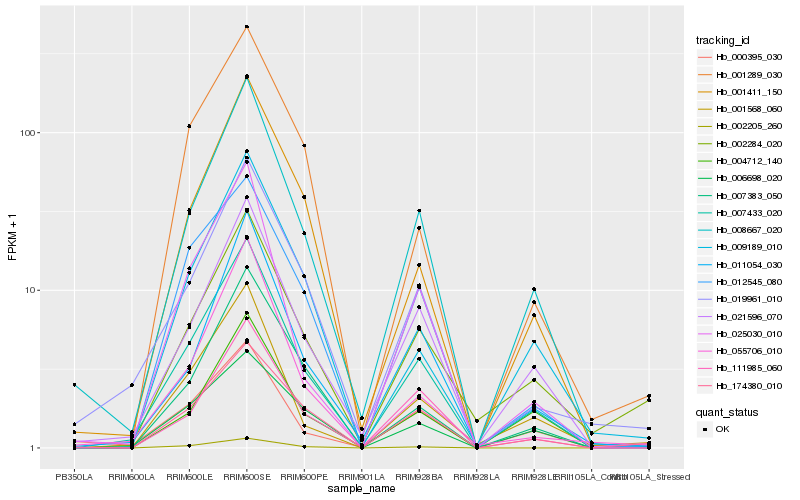
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007433_020 |
0.0 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 2 |
Hb_008667_020 |
0.0762929941 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_021596_070 |
0.0850270917 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_019961_010 |
0.1180161831 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 5 |
Hb_001411_150 |
0.1213051625 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 6 |
Hb_011054_030 |
0.121555093 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 7 |
Hb_111985_060 |
0.12594433 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 8 |
Hb_001568_060 |
0.1261467538 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 9 |
Hb_174380_010 |
0.1264573388 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_007383_050 |
0.1299978352 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 11 |
Hb_000395_030 |
0.1303474275 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 12 |
Hb_012545_080 |
0.1304156631 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 13 |
Hb_004712_140 |
0.131590371 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 14 |
Hb_025030_010 |
0.1315945869 |
- |
- |
- |
| 15 |
Hb_002284_020 |
0.1325576038 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 16 |
Hb_002205_260 |
0.1345718434 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 17 |
Hb_006698_020 |
0.1347409303 |
- |
- |
hypothetical protein JCGZ_04585 [Jatropha curcas] |
| 18 |
Hb_009189_010 |
0.1348832902 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 19 |
Hb_055706_010 |
0.1390961737 |
- |
- |
hypothetical protein JCGZ_22222 [Jatropha curcas] |
| 20 |
Hb_001289_030 |
0.1405057049 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |