| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
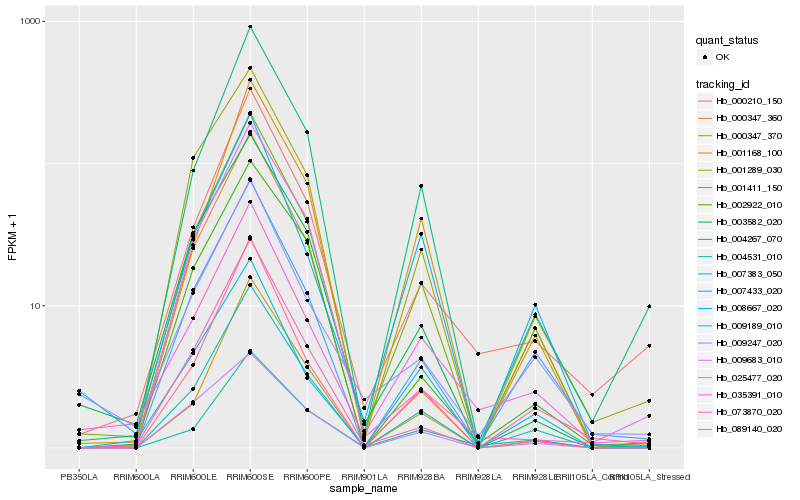
Hb_001411_150 |
0.0 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 2 |
Hb_007383_050 |
0.0397156539 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 3 |
Hb_009189_010 |
0.0766895106 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 4 |
Hb_001289_030 |
0.0906657372 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 5 |
Hb_004267_070 |
0.0953307712 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 6 |
Hb_001168_100 |
0.0971260802 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2-like [Citrus sinensis] |
| 7 |
Hb_089140_020 |
0.1036947869 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_003582_020 |
0.1041486438 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Eucalyptus grandis] |
| 9 |
Hb_025477_020 |
0.1042069048 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF027 [Jatropha curcas] |
| 10 |
Hb_073870_020 |
0.1060493364 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 11 |
Hb_000347_370 |
0.1075084838 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 12 |
Hb_000347_360 |
0.1110728451 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004531_010 |
0.1111664454 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |
| 14 |
Hb_000210_150 |
0.114461892 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 15 |
Hb_002922_010 |
0.1196353977 |
- |
- |
heavy-metal-associated domain-containing family protein [Populus trichocarpa] |
| 16 |
Hb_007433_020 |
0.1213051625 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 17 |
Hb_009683_010 |
0.1217295195 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_008667_020 |
0.122817305 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 19 |
Hb_009247_020 |
0.1235688739 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 20 |
Hb_035391_010 |
0.12384157 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |