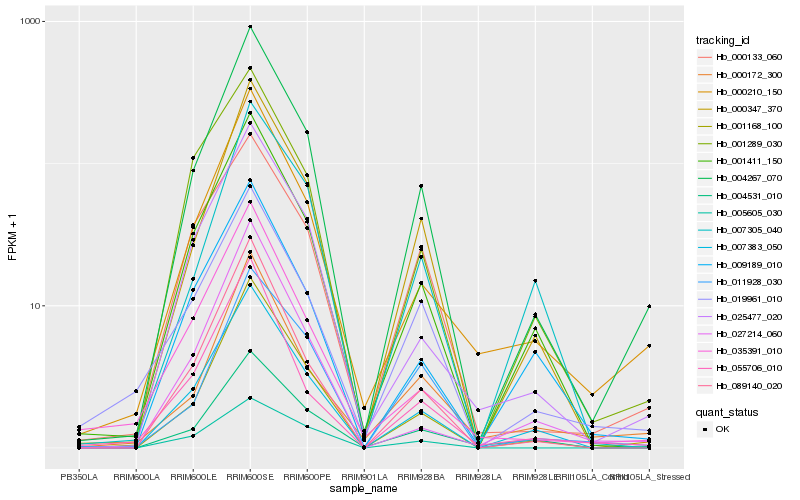
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004267_070 |
0.0 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 2 |
Hb_001168_100 |
0.0778158023 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2-like [Citrus sinensis] |
| 3 |
Hb_000347_370 |
0.0788572023 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 4 |
Hb_007383_050 |
0.0872302751 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 5 |
Hb_001411_150 |
0.0953307712 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 6 |
Hb_000210_150 |
0.0974804705 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 7 |
Hb_089140_020 |
0.100315396 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_004531_010 |
0.1068357186 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |
| 9 |
Hb_025477_020 |
0.1099056641 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF027 [Jatropha curcas] |
| 10 |
Hb_000172_300 |
0.1235543938 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 11 |
Hb_001289_030 |
0.1296183938 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 12 |
Hb_011928_030 |
0.1321898325 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 13 |
Hb_009189_010 |
0.1323671766 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 14 |
Hb_035391_010 |
0.1347483669 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 15 |
Hb_019961_010 |
0.134980422 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 16 |
Hb_000133_060 |
0.1353790978 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 17 |
Hb_055706_010 |
0.1355097333 |
- |
- |
hypothetical protein JCGZ_22222 [Jatropha curcas] |
| 18 |
Hb_007305_040 |
0.1360260077 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 19 |
Hb_027214_060 |
0.1361505148 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 20 |
Hb_005605_030 |
0.1376358776 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |