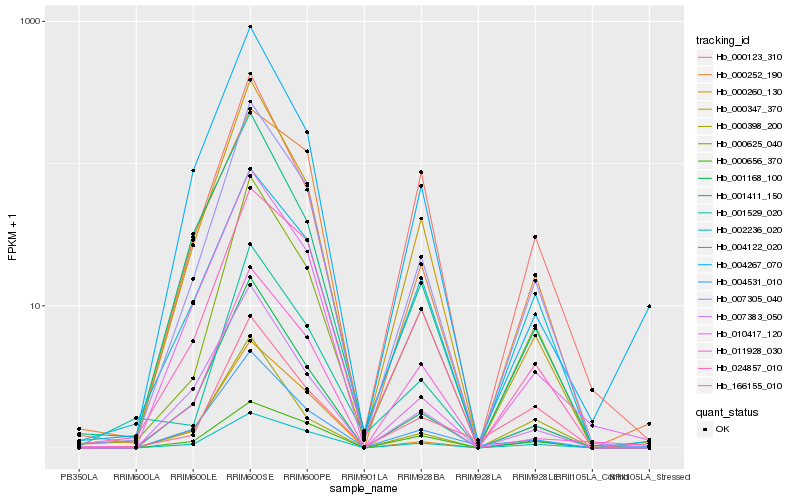
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007305_040 |
0.0 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 2 |
Hb_000625_040 |
0.0776995432 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 3 |
Hb_002236_020 |
0.089915764 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 4 |
Hb_000347_370 |
0.1007817344 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 5 |
Hb_024857_010 |
0.1016863385 |
- |
- |
glutaredoxin 2.1 [Hevea brasiliensis] |
| 6 |
Hb_004531_010 |
0.1070616513 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |
| 7 |
Hb_000260_130 |
0.113348503 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001168_100 |
0.1159812505 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2-like [Citrus sinensis] |
| 9 |
Hb_007383_050 |
0.1173695629 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 10 |
Hb_004122_020 |
0.126519865 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 11 |
Hb_001411_150 |
0.1312990259 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 12 |
Hb_011928_030 |
0.1339485634 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 13 |
Hb_004267_070 |
0.1360260077 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 14 |
Hb_000252_190 |
0.1367333664 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 15 |
Hb_000656_370 |
0.1400662865 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_001529_020 |
0.1407217524 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000398_200 |
0.1412279411 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 18 |
Hb_010417_120 |
0.1437395493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_166155_010 |
0.144368994 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 20 |
Hb_000123_310 |
0.1488821399 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |