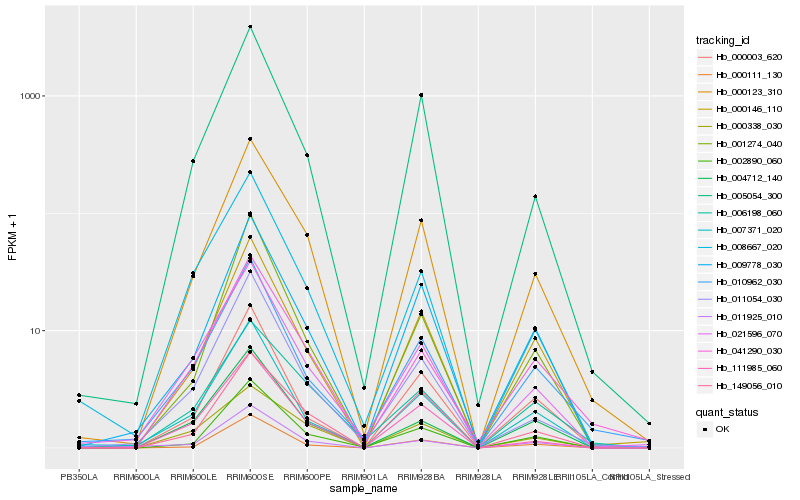
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000123_310 |
0.0 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 2 |
Hb_009778_030 |
0.0897820312 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 3 |
Hb_002890_060 |
0.09041572 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 4 |
Hb_000146_110 |
0.0933945634 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 5 |
Hb_005054_300 |
0.102461101 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 6 |
Hb_041290_030 |
0.1035657998 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 7 |
Hb_021596_070 |
0.1053400397 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_011054_030 |
0.1074269012 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 9 |
Hb_149056_010 |
0.1079418673 |
- |
- |
hypothetical protein CICLE_v10016664mg [Citrus clementina] |
| 10 |
Hb_111985_060 |
0.1097401168 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 11 |
Hb_006198_060 |
0.1104260104 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 12 |
Hb_001274_040 |
0.1146151257 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 13 |
Hb_000338_030 |
0.1174544508 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_21254 [Jatropha curcas] |
| 14 |
Hb_010962_030 |
0.1177124863 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_007371_020 |
0.1224581783 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000111_130 |
0.1246097456 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 17 |
Hb_008667_020 |
0.125200513 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_011925_010 |
0.1262242892 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000003_620 |
0.1269051435 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 20 |
Hb_004712_140 |
0.127389595 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |